Loss of PARP7 Increases Type I Interferon Signaling in EO771 Breast Cancer Cells and Prevents Mammary Tumor Growth by Increasing Antitumor Immunity
- PMID: 37509350
- PMCID: PMC10377955
- DOI: 10.3390/cancers15143689
Loss of PARP7 Increases Type I Interferon Signaling in EO771 Breast Cancer Cells and Prevents Mammary Tumor Growth by Increasing Antitumor Immunity
Abstract
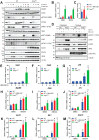
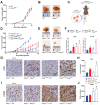
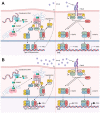
PARP7 is a member of the ADP-ribosyltransferase diphtheria toxin-like (ARTD) family and acts as a repressor of type I interferon (IFN) signaling. PARP7 inhibition causes tumor regression by enhancing antitumor immunity, which is dependent on the stimulator of interferon genes (STING) pathway, TANK-binding kinase 1 (TBK1) activity, and cytotoxic CD8+ T cells. To better understand PARP7's role in cancer, we generated and characterized PARP7 knockout (Parp7KO) EO771 mouse mammary cancer cells in vitro and in a preclinical syngeneic tumor model using catalytic mutant Parp7H532A mice. Loss of PARP7 expression or inhibition of its activity increased type I IFN signaling, as well as the levels of interferon-stimulated gene factor 3 (ISGF3) and specifically unphosphorylated-ISGF3 regulated target genes. This was partly because PARP7's modification of the RelA subunit of nuclear factor κ-B (NF-κB). PARP7 loss had no effect on tumor growth in immunodeficient mice. In contrast, injection of wildtype cells into Parp7H532A mice resulted in smaller tumors compared with cells injected into Parp7+/+ mice. Parp7H532A mice injected with Parp7KO cells failed to develop tumors and those that developed regressed. Our data highlight the importance of PARP7 in the immune cells and further support targeting PARP7 for anticancer therapy.
Keywords: ADP-ribosylation; PARP7; breast cancer; tumor immunity; type I interferon.
Conflict of interest statement
J.M. is a consultant for Duke Street Bio Inc.
Figures




Similar articles
-
Unleashing viral mimicry: A combinatorial strategy to enhance the efficacy of PARP7 inhibitors.Bioessays. 2025 Feb;47(2):e2400087. doi: 10.1002/bies.202400087. Epub 2024 Nov 6. Bioessays. 2025. PMID: 39502005 Free PMC article. Review.
-
PARP7 inhibits type I interferon signaling to prevent autoimmunity and lung disease.J Exp Med. 2025 May 5;222(5):e20241184. doi: 10.1084/jem.20241184. Epub 2025 Feb 19. J Exp Med. 2025. PMID: 39969510 Free PMC article.
-
Loss of Parp7 increases type I interferon signalling and reduces pancreatic tumour growth by enhancing immune cell infiltration.Front Immunol. 2025 Jan 10;15:1513595. doi: 10.3389/fimmu.2024.1513595. eCollection 2024. Front Immunol. 2025. PMID: 39867896 Free PMC article.
-
Structurally distinct PARP7 inhibitors provide new insights into the function of PARP7 in regulating nucleic acid-sensing and IFN-β signaling.Cell Chem Biol. 2023 Jan 19;30(1):43-54.e8. doi: 10.1016/j.chembiol.2022.11.012. Epub 2022 Dec 17. Cell Chem Biol. 2023. PMID: 36529140 Free PMC article.
-
PARP7 as a new target for activating anti-tumor immunity in cancer.EMBO Mol Med. 2025 May;17(5):872-888. doi: 10.1038/s44321-025-00214-6. Epub 2025 Mar 24. EMBO Mol Med. 2025. PMID: 40128585 Free PMC article. Review.
Cited by
-
Expanding the Perspective on PARP1 and Its Inhibitors in Cancer Therapy: From DNA Damage Repair to Immunomodulation.Biomedicines. 2024 Jul 20;12(7):1617. doi: 10.3390/biomedicines12071617. Biomedicines. 2024. PMID: 39062190 Free PMC article. Review.
-
PARP enzymes and mono-ADP-ribosylation: advancing the connection from interferon-signalling to cancer biology.Expert Rev Mol Med. 2024 Aug 27;26:e17. doi: 10.1017/erm.2024.13. Expert Rev Mol Med. 2024. PMID: 39189367 Free PMC article. Review.
-
Unleashing viral mimicry: A combinatorial strategy to enhance the efficacy of PARP7 inhibitors.Bioessays. 2025 Feb;47(2):e2400087. doi: 10.1002/bies.202400087. Epub 2024 Nov 6. Bioessays. 2025. PMID: 39502005 Free PMC article. Review.
-
PARP7 inhibits type I interferon signaling to prevent autoimmunity and lung disease.J Exp Med. 2025 May 5;222(5):e20241184. doi: 10.1084/jem.20241184. Epub 2025 Feb 19. J Exp Med. 2025. PMID: 39969510 Free PMC article.
-
A multilineage screen identifies actionable synthetic lethal interactions in human cancers.Nat Genet. 2025 Jan;57(1):154-164. doi: 10.1038/s41588-024-01971-9. Epub 2024 Nov 18. Nat Genet. 2025. PMID: 39558023
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

