Repurposing Anti-Dengue Compounds against Monkeypox Virus Targeting Core Cysteine Protease
- PMID: 37509664
- PMCID: PMC10377189
- DOI: 10.3390/biomedicines11072025
Repurposing Anti-Dengue Compounds against Monkeypox Virus Targeting Core Cysteine Protease
Abstract
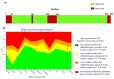
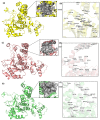
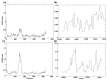
The monkeypox virus (MPXV) is an enveloped, double-stranded DNA virus belonging to the genus Orthopox viruses. In recent years, the virus has spread to countries where it was previously unknown, turning it into a worldwide emergency for public health. This study employs a structural-based drug design approach to identify potential inhibitors for the core cysteine proteinase of MPXV. During the simulations, the study identified two potential inhibitors, compound CHEMBL32926 and compound CHEMBL4861364, demonstrating strong binding affinities and drug-like properties. Their docking scores with the target protein were -10.7 and -10.9 kcal/mol, respectively. This study used ensemble-based protein-ligand docking to account for the binding site conformation variability. By examining how the identified inhibitors interact with the protein, this research sheds light on the workings of the inhibitors' mechanisms of action. Molecular dynamic simulations of protein-ligand complexes showed fluctuations from the initial docked pose, but they confirmed their binding throughout the simulation. The MMGBSA binding free energy calculations for CHEMBL32926 showed a binding free energy range of (-9.25 to -9.65) kcal/mol, while CHEMBL4861364 exhibited a range of (-41.66 to -31.47) kcal/mol. Later, analogues were searched for these compounds with 70% similarity criteria, and their IC50 was predicted using pre-trained machine learning models. This resulted in identifying two similar compounds for each hit with comparable binding affinity for cysteine proteinase. This study's structure-based drug design approach provides a promising strategy for identifying new drugs for treating MPXV infections.
Keywords: cysteine proteinase; in silico methods; molecular docking; molecular dynamic simulations; monkeypox virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Repositioning of anti-infective compounds against monkeypox virus core cysteine proteinase: a molecular dynamics study.Mol Divers. 2024 Dec;28(6):4113-4135. doi: 10.1007/s11030-023-10802-8. Epub 2024 Apr 23. Mol Divers. 2024. PMID: 38652365
-
Identifying potential monkeypox virus inhibitors: an in silico study targeting the A42R protein.Front Cell Infect Microbiol. 2024 Mar 4;14:1351737. doi: 10.3389/fcimb.2024.1351737. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38500508 Free PMC article.
-
Exploration of Microbially Derived Natural Compounds against Monkeypox Virus as Viral Core Cysteine Proteinase Inhibitors.Viruses. 2023 Jan 16;15(1):251. doi: 10.3390/v15010251. Viruses. 2023. PMID: 36680291 Free PMC article.
-
Bioprospecting of Meliaceae family phytomolecules for the treatment of monkeypox virus infection: a QSAR modeling and MD simulation approach.J Biomol Struct Dyn. 2025 Mar;43(5):2277-2299. doi: 10.1080/07391102.2023.2294180. Epub 2024 Jan 4. J Biomol Struct Dyn. 2025. PMID: 38174404
-
A chemoinformatic-biophysics based approach to identify novel anti-virulent compounds against Pseudomonas aeruginosa disulfide-bond protein A1.J Biomol Struct Dyn. 2023 Aug 7:1-10. doi: 10.1080/07391102.2023.2245470. Online ahead of print. J Biomol Struct Dyn. 2023. PMID: 37551016 Review.
Cited by
-
Revisiting methotrexate and phototrexate Zinc15 library-based derivatives using deep learning in-silico drug design approach.Front Chem. 2024 Mar 21;12:1380266. doi: 10.3389/fchem.2024.1380266. eCollection 2024. Front Chem. 2024. PMID: 38576849 Free PMC article.
-
Computational investigation of potential natural compounds as inhibitors of monkeypox virus cysteine proteinase.Front Bioinform. 2025 Jul 28;5:1637207. doi: 10.3389/fbinf.2025.1637207. eCollection 2025. Front Bioinform. 2025. PMID: 40791850 Free PMC article.
-
Repositioning of anti-infective compounds against monkeypox virus core cysteine proteinase: a molecular dynamics study.Mol Divers. 2024 Dec;28(6):4113-4135. doi: 10.1007/s11030-023-10802-8. Epub 2024 Apr 23. Mol Divers. 2024. PMID: 38652365
References
-
- Bennett J.E., Dolin R., Blaser M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases E-Book. Elsevier Health Sciences; Amsterdam, The Netherlands: 2019.
Grants and funding
LinkOut - more resources
Full Text Sources

