A Case of Class I 17p13.3 Microduplication Syndrome with Unilateral Hearing Loss
- PMID: 37510238
- PMCID: PMC10379727
- DOI: 10.3390/genes14071333
A Case of Class I 17p13.3 Microduplication Syndrome with Unilateral Hearing Loss
Abstract
17p13 is a chromosomal region characterized by genomic instability due to high gene density leading to multiple deletion and duplication events. 17p13.3 microduplication syndrome is a rare condition, reported only in 40 cases worldwide, which is found in the Miller-Dieker chromosomal region, presenting a wide range of phenotypic manifestations. Usually, the duplicated area is de novo and varies in size from 1.8 to 4.0 Mbp. Critical genes for this region are PAFAH1B1 (#601545), YWHAE (#605066), and CRK (#164762). 17p13.3 microduplication syndrome can be categorized into two classes (Class I and Class II) based on the genes that are present in the duplicated area, which lead to different phenotypes. In this report, we present a new case of Class I 17p13.3 microduplication syndrome that presents with unilateral sensorineural hearing loss. Oligonucleotide and SNP array comparative genomic hybridization (a-CGH) analysis revealed a duplication of approximately 121 Kbp on chromosome 17p13.3, which includes YWHAE and CRK genes. Whole-exome sequencing (WES) analysis confirmed the duplication. Our patient has common clinical symptoms of Class I 17p13.3 microduplication syndrome, and in addition, she has unilateral sensorineural hearing loss. Interestingly, WES analysis did not detect any mutations in genes that are associated with hearing loss. The above findings lead us to propose that hearing loss is a manifestation of 17p13.3 duplication syndrome.
Keywords: 17p13.3; WES; YHWAE; array-CGH; microduplication; neurodevelopmental disorders.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures
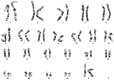


References
-
- Chen C.P., Chang T.Y., Guo W.Y., Wu P.C., Wang L.K., Chern S.R., Wu P.S., Su J.W., Chen Y.T., Chen L.F., et al. Chromosome 17p13.3 deletion syndrome: aCGH characterization, prenatal findings and diagnosis, and literature review. Gene. 2013;532:152–159. doi: 10.1016/j.gene.2013.09.044. - DOI - PubMed
-
- Bruno D.L., Anderlid B.-M., Lindstrand A., van Ravenswaaij-Arts C., Ganesamoorthy D., Lundin J., Martin C.L., Douglas J., Nowak C., Adam M.P., et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J. Med Genet. 2010;47:299–311. doi: 10.1136/jmg.2009.069906. - DOI - PubMed
-
- Armour C.M., E Bulman D., Jarinova O., Rogers R.C., Clarkson K.B., DuPont B.R., Dwivedi A., O Bartel F., McDonell L., E Schwartz C., et al. 17p13.3 microduplications are associated with split-hand/foot malformation and long-bone deficiency (SHFLD) Eur. J. Hum. Genet. 2011;19:1144–1151. doi: 10.1038/ejhg.2011.97. - DOI - PMC - PubMed
-
- Curry C.J., Rosenfeld J.A., Grant E., Gripp K.W., Anderson C., Aylsworth A.S., Ben Saad T., Chizhikov V.V., Dybose G., Fagerberg C., et al. The duplication 17p13.3 phenotype: Analysis of 21 families delineates developmental, behavioral and brain abnormalities, and rare variant phenotypes. Am. J. Med Genet. Part A. 2013;161A:1833–1852. doi: 10.1002/ajmg.a.35996. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

