Characterization and Potential Function Analysis of the SRS Gene Family in Brassica napus
- PMID: 37510325
- PMCID: PMC10379590
- DOI: 10.3390/genes14071421
Characterization and Potential Function Analysis of the SRS Gene Family in Brassica napus
Abstract
SRS (SHI-related sequence) transcription factors play a crucial role in plant growth, development, and abiotic stress response. Although Brassica napus (B. napus) is one of the most important oil crops in the world, the role of SRS genes in B. napus (BnSRS) has not been well investigated. Therefore, we employed a bioinformatics approach to identify BnSRS genes from genomic data and investigated their characteristics, functions, and expression patterns, to gain a better understanding of how this gene family is involved in plant development and growth. The results revealed that there were 34 BnSRS gene family members in the genomic sequence of B. napus, unevenly distributed throughout the sequence. Based on the phylogenetic analysis, these BnSRS genes could be divided into four subgroups, with each group sharing comparable conserved motifs and gene structure. Analysis of the upstream promoter region showed that BnSRS genes may regulate hormone responses, biotic and abiotic stress response, growth, and development in B. napus. The protein-protein interaction analysis revealed the involvement of BnSRS genes in various biological processes and metabolic pathways. Our analysis of BnSRS gene expression showed that 23 BnSRS genes in the callus tissue exhibited a dominant expression pattern, suggesting their critical involvement in cell dedifferentiation, cell division, and tissue development. In addition, association analysis between genotype and agronomic traits revealed that BnSRS genes may be linked to some important agronomic traits in B. napus, suggesting that BnSRS genes were widely involved in the regulation of important agronomic traits (including C16.0, C18.0, C18.1, C18.2 C18.3, C20.1, C22.1, GLU, protein, TSW, and FFT). In this study, we predicted the evolutionary relationships and potential functions of BnSRS gene family members, providing a basis for the development of BnSRS gene functions which could facilitate targeted functional studies and genetic improvement for elite breeding in B. napus.
Keywords: Brassica napus; SRS gene family; agronomic traits; expression pattern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
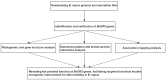




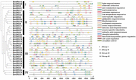




References
-
- Eklund D.M., Cierlik I., Ståldal V., Claes A.R., Vestman D., Chandler J., Sundberg E. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 2011;157:2069–2080. doi: 10.1104/pp.111.182253. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

