High-Resolution Genotyping of Expressed Equine MHC Reveals a Highly Complex MHC Structure
- PMID: 37510326
- PMCID: PMC10379315
- DOI: 10.3390/genes14071422
High-Resolution Genotyping of Expressed Equine MHC Reveals a Highly Complex MHC Structure
Abstract
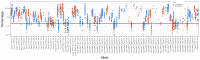
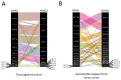
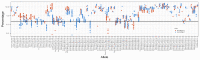
The Major Histocompatibility Complex (MHC) genes play a key role in a number of biological processes, most notably in immunological responses. The MHCI and MHCII genes incorporate a complex set of highly polymorphic and polygenic series of genes, which, due to the technical limitations of previously available technologies, have only been partially characterized in non-model but economically important species such as the horse. The advent of high-throughput sequencing platforms has provided new opportunities to develop methods to generate high-resolution sequencing data on a large scale and apply them to the analysis of complex gene sets such as the MHC. In this study, we developed and applied a MiSeq-based approach for the combined analysis of the expressed MHCI and MHCII repertoires in cohorts of Thoroughbred, Icelandic, and Norwegian Fjord Horses. The approach enabled us to generate comprehensive MHCI/II data for all of the individuals (n = 168) included in the study, identifying 152 and 117 novel MHCI and MHCII sequences, respectively. There was limited overlap in MHCI and MHCII haplotypes between the Thoroughbred and the Icelandic/Norwegian Fjord horses, showcasing the variation in MHC repertoire between genetically divergent breeds, and it can be inferred that there is much more MHC diversity in the global horse population. This study provided novel insights into the structure of the expressed equine MHC repertoire and highlighted unique features of the MHC in horses.
Keywords: Major Histocompatibility Complex (MHC); breed diversity; equine; high-throughput sequencing; polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Rapid identification of bovine MHCI haplotypes in genetically divergent cattle populations using next-generation sequencing.Immunogenetics. 2016 Nov;68(10):765-781. doi: 10.1007/s00251-016-0945-7. Epub 2016 Aug 11. Immunogenetics. 2016. PMID: 27516207 Free PMC article.
-
High throughput analysis of MHC-I and MHC-DR diversity of Brazilian cattle populations.HLA. 2021 Aug;98(2):93-113. doi: 10.1111/tan.14339. Epub 2021 Jun 17. HLA. 2021. PMID: 34102036
-
High throughput analysis of MHC class I and class II diversity of Zambian indigenous cattle populations.HLA. 2023 May;101(5):458-483. doi: 10.1111/tan.14976. Epub 2023 Jan 29. HLA. 2023. PMID: 36680506 Free PMC article.
-
The cattle major histocompatibility complex: is it unique?Vet Immunol Immunopathol. 2004 Nov;102(1-2):1-8. doi: 10.1016/j.vetimm.2004.06.007. Vet Immunol Immunopathol. 2004. PMID: 15451610 Review.
-
Building proteomic tool boxes to monitor MHC class I and class II peptides.Proteomics. 2017 Jan;17(1-2). doi: 10.1002/pmic.201600061. Proteomics. 2017. PMID: 27928884 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

