SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia
- PMID: 37511096
- PMCID: PMC10379212
- DOI: 10.3390/ijms241411335
SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia
Abstract
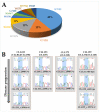
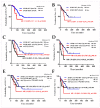
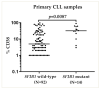
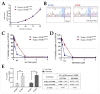
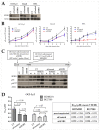
Chronic lymphocytic leukemia (CLL) is a genetically and clinically heterogeneous malignancy affecting older individuals. There are a number of current treatment options for CLL, including monoclonal antibodies, targeted drugs, chemotherapy, and different combinations of these. However, for those patients who are intrinsically treatment resistant, or relapse following initial responses, novel targeted therapies are still needed. Targeting the mouse double-minute-2 human homolog (MDM2), a primary negative regulator of p53, is an appealing therapeutic strategy for non-genotoxic reactivation of p53, since the TP53 gene is in its wild-type state at diagnosis in approximately 90% of patients. Mutated SF3B1 and TP53 are both associated with more aggressive disease, resistance to therapies and poorer overall survival for CLL. In this study, we performed a screen for SF3B1 and TP53 mutations and tested RG7388 (idasanutlin), a second-generation MDM2 inhibitor, in a cohort of CLL primary patient samples. SF3B1 mutations were detected in 24 of 195 cases (12.3%) and found associated with poor overall survival (hazard ratio [HR] 2.12, p = 0.032) and high CD38 expression (median CD38 (%) 32 vs. 5; p = 0.0087). The novel striking finding of this study was an independent link between SF3B1 mutational status and poor response to RG7388. Overall, SF3B1 mutations in CLL patient samples were associated with resistance to treatment with RG7388 ex vivo, and patients with the wild type for both SF3B1 and TP53 are more likely to benefit from treatment with MDM2 inhibitors.
Keywords: MDM2-p53 antagonists; RG7388 (idasanutlin); SF3B1; chronic lymphocytic leukemia (CLL); splicing factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ahn I.E., Tian X., Ipe D., Cheng M., Albitar M., Tsao L.C., Zhang L., Ma W., Herman S.E.M., Gaglione E.M., et al. Prediction of Outcome in Patients With Chronic Lymphocytic Leukemia Treated With Ibrutinib: Development and Validation of a Four-Factor Prognostic Model. J. Clin. Oncol. 2021;39:576–585. doi: 10.1200/JCO.20.00979. - DOI - PMC - PubMed
-
- Byrd J.C., Wierda W.G., Schuh A., Devereux S., Chaves J.M., Brown J.R., Hillmen P., Martin P., Awan F.T., Stephens D.M., et al. Acalabrutinib monotherapy in patients with relapsed/refractory chronic lymphocytic leukemia: Updated phase 2 results. Blood. 2020;135:1204–1213. doi: 10.1182/blood.2018884940. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

