Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects?
- PMID: 37511197
- PMCID: PMC10380557
- DOI: 10.3390/ijms241411437
Genomic Instability Evolutionary Footprints on Human Health: Driving Forces or Side Effects?
Abstract
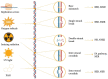
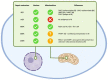
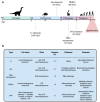
In this work, we propose a comprehensive perspective on genomic instability comprising not only the accumulation of mutations but also telomeric shortening, epigenetic alterations and other mechanisms that could contribute to genomic information conservation or corruption. First, we present mechanisms playing a role in genomic instability across the kingdoms of life. Then, we explore the impact of genomic instability on the human being across its evolutionary history and on present-day human health, with a particular focus on aging and complex disorders. Finally, we discuss the role of non-coding RNAs, highlighting future approaches for a better living and an expanded healthy lifespan.
Keywords: DNA repair; aging; evolutionary genetics; genomic instability; human complex disorder; ncRNA; neurodegenerative diseases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

