A Proteomic Survey of the Cystic Fibrosis Transmembrane Conductance Regulator Surfaceome
- PMID: 37511222
- PMCID: PMC10380767
- DOI: 10.3390/ijms241411457
A Proteomic Survey of the Cystic Fibrosis Transmembrane Conductance Regulator Surfaceome
Abstract
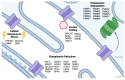
The aim of this review article is to collate recent contributions of proteomic studies to cystic fibrosis transmembrane conductance regulator (CFTR) biology. We summarize advances from these studies and create an accessible resource for future CFTR proteomic efforts. We focus our attention on the CFTR interaction network at the cell surface, thus generating a CFTR 'surfaceome'. We review the main findings about CFTR interactions and highlight several functional categories amongst these that could lead to the discovery of potential biomarkers and drug targets for CF.
Keywords: BioID; CFTR interactions; CFTR modulators; PDZ domain; clathrin-mediated endocytosis; cystic fibrosis; interactome; peripheral quality control; surfaceome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Deriy L.V., Gomez E.A., Zhang G., Beacham D.W., Hopson J.A., Gallan A.J., Shevchenko P.D., Bindokas V.P., Nelson D.J. Disease-causing Mutations in the Cystic Fibrosis Transmembrane Conductance Regulator Determine the Functional Responses of Alveolar Macrophages. J. Biol. Chem. 2009;284:35926–35938. doi: 10.1074/jbc.M109.057372. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

