Cell-Based Measurement of Mitochondrial Function in Human Airway Smooth Muscle Cells
- PMID: 37511264
- PMCID: PMC10380259
- DOI: 10.3390/ijms241411506
Cell-Based Measurement of Mitochondrial Function in Human Airway Smooth Muscle Cells
Abstract
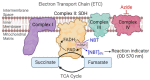
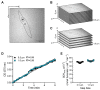
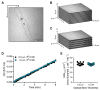
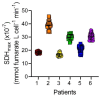
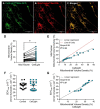
Cellular mitochondrial function can be assessed using high-resolution respirometry that measures the O2 consumption rate (OCR) across a number of cells. However, a direct measurement of cellular mitochondrial function provides valuable information and physiological insight. In the present study, we used a quantitative histochemical technique to measure the activity of succinate dehydrogenase (SDH), a key enzyme located in the inner mitochondrial membrane, which participates in both the tricarboxylic acid (TCA) cycle and electron transport chain (ETC) as Complex II. In this study, we determine the maximum velocity of the SDH reaction (SDHmax) in individual human airway smooth muscle (hASM) cells. To measure SDHmax, hASM cells were exposed to a solution containing 80 mM succinate and 1.5 mM nitroblue tetrazolium (NBT, reaction indicator). As the reaction proceeded, the change in optical density (OD) due to the reduction of NBT to its diformazan (peak absorbance wavelength of 570 nm) was measured using a confocal microscope with the pathlength for light absorbance tightly controlled. SDHmax was determined during the linear period of the SDH reaction and expressed as mmol fumarate/liter of cell/min. We determine that this technique is rigorous and reproducible, and reliable for the measurement of mitochondrial function in individual cells.
Keywords: airway smooth muscle; confocal microscope; mitochondria; oxygen consumption; succinate dehydrogenase.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

