Epigenetic Modulation of Inflammatory Pathways in Myometrial Stem Cells and Risk of Uterine Fibroids
- PMID: 37511399
- PMCID: PMC10380326
- DOI: 10.3390/ijms241411641
Epigenetic Modulation of Inflammatory Pathways in Myometrial Stem Cells and Risk of Uterine Fibroids
Abstract
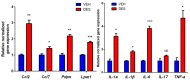
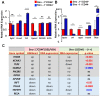
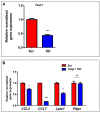
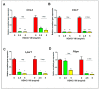
The period during which tissue and organ development occurs is particularly vulnerable to the influence of environmental exposures. However, the specific mechanisms through which biological pathways are disrupted in response to developmental insults, consequently elevating the risk of hormone-dependent diseases, such as uterine fibroids (UFs), remain poorly understood. Here, we show that developmental exposure to the endocrine-disrupting chemical (EDC), diethylstilbestrol (DES), activates the inflammatory pathways in myometrial stem cells (MMSCs), which are the origin of UFs. Significantly, the secretome of reprogrammed MMSCs enhances the expression of critical inflammation-related genes in differentiated myometrial cells through the paracrine mechanism, which amplifies pro-inflammatory and immune suppression signaling in the myometrium. The expression of reprogrammed inflammatory responsive genes (IRGs) is driven by activated mixed-lineage leukemia protein-1 (MLL1) in MMSCs. The deactivation of MLL reverses the reprogramming of IRG expression. In addition, the inhibition of histone deacetylases (HDACs) also reversed the reprogrammed IRG expression induced by EDC exposure. This work identifies the epigenetic mechanisms of MLL1/HDAC-mediated MMSC reprogramming, and EDC exposure epigenetically targets MMSCs and imparts an IRG expression pattern, which may result in a "hyper-inflammatory phenotype" and an increased hormone-dependent risk of UFs later in life.
Keywords: developmental reprogramming; diethylstilbestrol; inflammatory responsive genes; myometrial stem cells; uterine fibroids.
Conflict of interest statement
None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
Figures



References
-
- Segars J.H., Parrott E.C., Nagel J.D., Guo X.C., Gao X., Birnbaum L.S., Pinn V.W., Dixon D. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: Comprehensive review, conference summary and future recommendations. Hum. Reprod. Update. 2014;20:309–333. doi: 10.1093/humupd/dmt058. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

