A Severe Acute Respiratory Syndrome Coronavirus 2 Anti-Spike Immunoglobulin G Assay: A Robust Method for Evaluation of Vaccine Immunogenicity Using an Established Correlate of Protection
- PMID: 37512961
- PMCID: PMC10383018
- DOI: 10.3390/microorganisms11071789
A Severe Acute Respiratory Syndrome Coronavirus 2 Anti-Spike Immunoglobulin G Assay: A Robust Method for Evaluation of Vaccine Immunogenicity Using an Established Correlate of Protection
Abstract
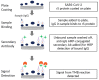
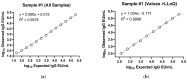
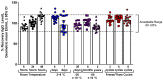
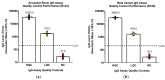
As the COVID-19 pandemic continues, variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to emerge. Immunogenicity evaluation of vaccines and identification of correlates of protection for vaccine effectiveness is critical to aid the development of vaccines against emerging variants. Anti-recombinant spike (rS) protein immunoglobulin G (IgG) quantitation in the systemic circulation (serum/plasma) is shown to correlate with vaccine efficacy. Thus, an enzyme-linked immunosorbent assay (ELISA)-based binding assay to detect SARS-CoV-2 (ancestral and variant strains) anti-rS IgG in human serum samples was developed and validated. This assay successfully met acceptance criteria for inter/intra-assay precision, specificity, selectivity, linearity, lower/upper limits of quantitation, matrix effects, and assay robustness. The analyte in serum was stable for up to 8 freeze/thaw cycles and 2 years in -80 °C storage. Similar results were observed for the Beta, Delta, and Omicron BA.1/BA.5/XBB.1.5 variant-adapted assays. Anti-rS IgG assay results correlated significantly with neutralization and receptor binding inhibition assays. In addition, usage of international reference standards allows data extrapolation to WHO international units (BAU/mL), facilitating comparison of results with other IgG assays. This anti-rS IgG assay is a robust, high-throughput method to evaluate binding IgG responses to S protein in serum, enabling rapid development of effective vaccines against emerging COVID-19 variants.
Keywords: Binding antibodies; COVID-19; ELISA; IgG; Omicron BA.1/BA.5/XBB.1.5 SARS-CoV-2 variants; antibodies; correlate of protection; variants of concern.
Conflict of interest statement
All the authors are employees and stockholders of Novavax, Inc.
Figures


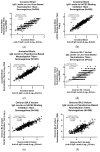
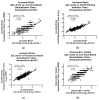

References
-
- Centers for Disease Control and Prevention David J. Spencer CDC Museum—COVID-19 Timeline. [(accessed on 3 February 2023)]; Available online: https://www.cdc.gov/museum/timeline/covid19.html.
-
- Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

