Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives
- PMID: 37512997
- PMCID: PMC10384668
- DOI: 10.3390/microorganisms11071825
Microbial-Derived Tryptophan Metabolites and Their Role in Neurological Disease: Anthranilic Acid and Anthranilic Acid Derivatives
Abstract
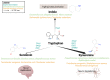
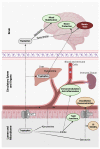
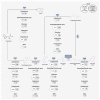
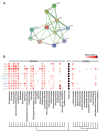
The gut microbiome provides the host access to otherwise indigestible nutrients, which are often further metabolized by the microbiome into bioactive components. The gut microbiome can also shift the balance of host-produced compounds, which may alter host health. One precursor to bioactive metabolites is the essential aromatic amino acid tryptophan. Tryptophan is mostly shunted into the kynurenine pathway but is also the primary metabolite for serotonin production and the bacterial indole pathway. Balance between tryptophan-derived bioactive metabolites is crucial for neurological homeostasis and metabolic imbalance can trigger or exacerbate neurological diseases. Alzheimer's, depression, and schizophrenia have been linked to diverging levels of tryptophan-derived anthranilic, kynurenic, and quinolinic acid. Anthranilic acid from collective microbiome metabolism plays a complex but important role in systemic host health. Although anthranilic acid and its metabolic products are of great importance for host-microbe interaction in neurological health, literature examining the mechanistic relationships between microbial production, host regulation, and neurological diseases is scarce and at times conflicting. This narrative review provides an overview of the current understanding of anthranilic acid's role in neurological health and disease, with particular focus on the contribution of the gut microbiome, the gut-brain axis, and the involvement of the three major tryptophan pathways.
Keywords: anthranilic acid; gut–brain axis; kynurenine; metabolites; microbiome; neuroactive compounds; tryptophan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures