Optimizing Crop Production with Bacterial Inputs: Insights into Chemical Dialogue between Sphingomonas sediminicola and Pisum sativum
- PMID: 37513019
- PMCID: PMC10385058
- DOI: 10.3390/microorganisms11071847
Optimizing Crop Production with Bacterial Inputs: Insights into Chemical Dialogue between Sphingomonas sediminicola and Pisum sativum
Abstract
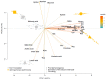
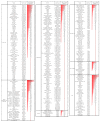
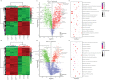
The use of biological inputs is an interesting approach to optimize crop production and reduce the use of chemical inputs. Understanding the chemical communication between bacteria and plants is critical to optimizing this approach. Recently, we have shown that Sphingomonas (S.) sediminicola can improve both nitrogen supply and yield in pea. Here, we used biochemical methods and untargeted metabolomics to investigate the chemical dialog between S. sediminicola and pea. We also evaluated the metabolic capacities of S. sediminicola by metabolic profiling. Our results showed that peas release a wide range of hexoses, organic acids, and amino acids during their development, which can generally recruit and select fast-growing organisms. In the presence of S. sediminicola, a more specific pattern of these molecules took place, gradually adapting to the metabolic capabilities of the bacterium, especially for pentoses and flavonoids. In turn, S. sediminicola is able to produce several compounds involved in cell differentiation, biofilm formation, and quorum sensing to shape its environment, as well as several molecules that stimulate pea growth and plant defense mechanisms.
Keywords: Sphingomonas sediminicola; molecular dialogue plant–bacteria; pea; plant–bacteria interaction; root exudates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kumari B., Mallick M.A., Solanki M.K., Solanki A.C., Hora A., Guo W. Plant Growth Promoting Rhizobacteria (PGPR): Modern Prospects for Sustainable Agriculture. In: Ansari R.A., Mahmood I., editors. Plant Health Under Biotic Stress. Springer; Singapore: 2019. pp. 109–127. Volume 2: Microbial Interactions.
-
- Oleńska E., Małek W., Wójcik M., Swiecicka I., Thijs S., Vangronsveld J. Beneficial Features of Plant Growth-Promoting Rhizobacteria for Improving Plant Growth and Health in Challenging Conditions: A Methodical Review. Sci. Total Environ. 2020;743:140682. doi: 10.1016/j.scitotenv.2020.140682. - DOI - PubMed
-
- Khan A.L., Waqas M., Kang S.-M., Al-Harrasi A., Hussain J., Al-Rawahi A., Al-Khiziri S., Ullah I., Ali L., Jung H.-Y., et al. Bacterial Endophyte Sphingomonas Sp. LK11 Produces Gibberellins and IAA and Promotes Tomato Plant Growth. J. Microbiol. 2014;52:689–695. doi: 10.1007/s12275-014-4002-7. - DOI - PubMed
-
- Etesami H., Adl S.M. Plant Growth-Promoting Rhizobacteria (PGPR) and Their Action Mechanisms in Availability of Nutrients to Plants. In: Kumar M., Kumar V., Prasad R., editors. Phyto-Microbiome in Stress Regulation. Springer; Singapore: 2020. pp. 147–203. Environmental and Microbial Biotechnology.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

