qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer
- PMID: 37514259
- PMCID: PMC10385359
- DOI: 10.3390/plants12142645
qPCR Assay as a Tool for Examining Cotton Resistance to the Virus Complex Causing CLCuD: Yield Loss Inversely Correlates with Betasatellite, Not Virus, DNA Titer
Abstract
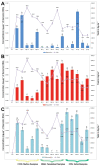
Cotton leaf curl disease (CLCuD) is a significant constraint to the economies of Pakistan and India. The disease is caused by different begomoviruses (genus Begomovirus, family Geminiviridae) in association with a disease-specific betasatellite. However, another satellite-like molecule, alphasatellite, is occasionally found associated with this disease complex. A quantitative real-time PCR assay for the virus/satellite components causing CLCuD was used to investigate the performance of selected cotton varieties in the 2014-2015 National Coordinated Varietal Trials (NCVT) in Pakistan. The DNA levels of virus and satellites in cotton plants were determined for five cotton varieties across three geographic locations and compared with seed cotton yield (SCY) as a measure of the plant performance. The highest virus titer was detected in B-10 (0.972 ng·µg-1) from Vehari and the lowest in B-3 (0.006 ng·µg-1) from Faisalabad. Likewise, the highest alphasatellite titer was found in B-1 (0.055 ng·µg-1) from Vehari and the lowest in B-1 and B-2 (0.001 ng·µg-1) from Faisalabad. The highest betasatellite titer was found in B-23 (1.156 ng·µg-1) from Faisalabad and the lowest in B-12 (0.072 ng·µg-1) from Multan. Virus/satellite DNA levels, symptoms, and SCY were found to be highly variable between the varieties and between the locations. Nevertheless, statistical analysis of the results suggested that betasatellite DNA levels, rather than virus or alphasatellite DNA levels, were the important variable in plant performance, having an inverse relationship with SCY (-0.447). This quantitative assay will be useful in breeding programs for development of virus resistant plants and varietal trials, such as the NCVT, to select suitable varieties of cotton with mild (preferably no) symptoms and low (preferably no) virus/satellite. At present, no such molecular techniques are used in resistance breeding programs or varietal trials in Pakistan.
Keywords: cotton leaf curl Khokhran virus—Burewala strain (CLCuKoV-Bur); cotton leaf curl Multan betasatellite (CLCuMuB); cotton leaf curl disease (CLCuD); national coordinated varietal trials (NCVT); quantitative real-time PCR (qPCR); seed cotton yield (SCY).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Real-time quantitative PCR assay for the quantification of virus and satellites causing leaf curl disease in cotton in Pakistan.J Virol Methods. 2017 Oct;248:54-60. doi: 10.1016/j.jviromet.2017.05.012. Epub 2017 May 29. J Virol Methods. 2017. PMID: 28572041
-
First Report of Okra enation leaf curl virus and Associated Cotton leaf curl Multan betasatellite and Cotton leaf curl Multan alphasatellite Infecting Cotton in Pakistan: A New Member of the Cotton Leaf Curl Disease Complex.Plant Dis. 2014 Oct;98(10):1447. doi: 10.1094/PDIS-04-14-0345-PDN. Plant Dis. 2014. PMID: 30704006
-
First Detection of Cotton leaf curl Burewala virus and Cognate Cotton leaf curl Multan betasatellite and Gossypium darwinii symptomless alphasatellite in Symptomatic Luffa cylindrica in Pakistan.Plant Dis. 2013 Aug;97(8):1122. doi: 10.1094/PDIS-12-12-1159-PDN. Plant Dis. 2013. PMID: 30722479
-
An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex.Viruses. 2017 Sep 29;9(10):280. doi: 10.3390/v9100280. Viruses. 2017. PMID: 28961220 Free PMC article. Review.
-
Cotton leaf curl disease - an emerging threat to cotton production worldwide.J Gen Virol. 2013 Apr;94(Pt 4):695-710. doi: 10.1099/vir.0.049627-0. Epub 2013 Jan 16. J Gen Virol. 2013. PMID: 23324471 Review.
Cited by
-
Comparative analysis, diversification, and functional validation of plant nucleotide-binding site domain genes.Sci Rep. 2024 May 24;14(1):11930. doi: 10.1038/s41598-024-62876-5. Sci Rep. 2024. PMID: 38789717 Free PMC article.
-
The Role of Satellites in the Evolution of Begomoviruses.Viruses. 2024 Jun 17;16(6):970. doi: 10.3390/v16060970. Viruses. 2024. PMID: 38932261 Free PMC article. Review.
-
Temporal changes in the levels of virus and betasatellite DNA in B. tabaci feeding on CLCuD affected cotton during the growing season.Front Microbiol. 2024 May 22;15:1410568. doi: 10.3389/fmicb.2024.1410568. eCollection 2024. Front Microbiol. 2024. PMID: 38841073 Free PMC article.
-
Differential interactions of ToLCNDV with different betasatellites reveal complex viral dynamics in N. benthamiana.PLoS One. 2025 Jun 25;20(6):e0327234. doi: 10.1371/journal.pone.0327234. eCollection 2025. PLoS One. 2025. PMID: 40561144 Free PMC article.
-
A comprehensive review on Gossypium hirsutum resistance against cotton leaf curl virus.Front Genet. 2024 Feb 19;15:1306469. doi: 10.3389/fgene.2024.1306469. eCollection 2024. Front Genet. 2024. PMID: 38440193 Free PMC article. Review.
References
-
- Ur-Rahman M., Rahmat Z., Mahmood A., Abdullah K., Zafar Y. Cotton Germplasm of Pakistan. In: Ibrokhim Y.A., editor. World Cotton Germplasm Resources. IntechOpen; Rijeka, Croatia: 2014. p. Ch. 6.
-
- FAOStat Food and Agriculture Organization of the United Nations Statistics Division. [(accessed on 25 April 2023)]. Available online: http://faostat3.fao.org/home/E.
-
- Briddon R.W., Bull S.E., Amin I., Mansoor S., Bedford I.D., Rishi N., Siwatch S.S., Zafar M.Y., Abdel-Salam A.M., Markham P.G. Diversity of DNA 1; a satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology. 2004;324:462–474. doi: 10.1016/j.virol.2004.03.041. - DOI - PubMed
-
- Hussain T., Ali M. A review of cotton diseases of Pakistan. Pak. Cotton. 1975;19:71–86.
LinkOut - more resources
Full Text Sources

