Rapid Generation of Recombinant Flaviviruses Using Circular Polymerase Extension Reaction
- PMID: 37515065
- PMCID: PMC10383701
- DOI: 10.3390/vaccines11071250
Rapid Generation of Recombinant Flaviviruses Using Circular Polymerase Extension Reaction
Abstract
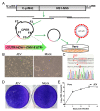
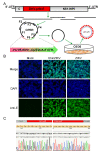
The genus Flavivirus is a group of arthropod-borne single-stranded RNA viruses, which includes important human and animal pathogens such as Japanese encephalitis virus (JEV), Zika virus (ZIKV), Dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), and Tick-borne encephalitis virus (TBEV). Reverse genetics has been a useful tool for understanding biological properties and the pathogenesis of flaviviruses. However, the conventional construction of full-length infectious clones for flavivirus is time-consuming and difficult due to the toxicity of the flavivirus genome to E. coli. Herein, we applied a simple, rapid, and bacterium-free circular polymerase extension reaction (CPER) method to synthesize recombinant flaviviruses in vertebrate cells as well as insect cells. We started with the de novo synthesis of the JEV vaccine strain SA-14-14-2 in Vero cells using CPER, and then modified the CPER method to recover insect-specific flaviviruses (ISFs) in mosquito C6/36 cells. Chimeric Zika virus (ChinZIKV) based on the Chaoyang virus (CYV) backbone and the Culex flavivirus reporter virus expressing green fluorescent protein (CxFV-GFP) were subsequently rescued in C6/36 cells. CPER is a simple method for the rapid generation of flaviviruses and other potential RNA viruses. A CPER-based recovery system for flaviviruses of different host ranges was established, which would facilitate the development of countermeasures against flavivirus outbreaks in the future.
Keywords: CPER; Flavivirus; reporter virus; reverse genetic; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Government of Canada Surveillance of West Nile Virus. [(accessed on 19 May 2023)]; Available online: https://www.canada.ca/en/public-health/services/diseases/west-nile-virus....
Grants and funding
LinkOut - more resources
Full Text Sources

