Structure of Bovine CD46 Ectodomain
- PMID: 37515111
- PMCID: PMC10385506
- DOI: 10.3390/v15071424
Structure of Bovine CD46 Ectodomain
Abstract
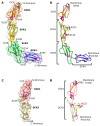
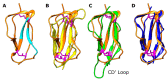
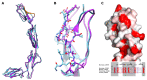
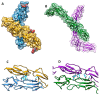
CD46, or membrane cofactor protein, is a type-one transmembrane protein from the complement regulatory protein family. Alongside its role in complement activation, CD46 is involved in many other processes, from T-cell activation to reproduction. It is also referred to as a pathogen magnet, because it is used as a receptor by multiple bacteria and viruses. Bovine CD46 (bovCD46) in particular is involved in bovine viral diarrhoea virus entry, an economically important disease in cattle industries. This study presents the X-ray crystallographic structure of the extracellular region of bovCD46, revealing a four-short-consensus-repeat (SCR) structure similar to that in human CD46. SCR1-3 are arranged linearly, while SCR 4 has a reduced interface angle, resulting in a hockey stick-like appearance. The structure also reveals the bovine viral diarrhoea virus interaction site in SCR1, which is likely to confer pestivirus specificity for their target host, CD46. Insights gained from the structural information on pestivirus receptors, such as CD46, could offer valuable guidance for future control strategies.
Keywords: BVDV; CD46; X-ray structure; bovine viral diarrhoea virus; pestivirus; type I membrane protein; virus receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Persson B.D., Schmitz N.B., Santiago C., Zocher G., Larvie M., Scheu U., Casasnovas J.M., Stehle T. Structure of the Extracellular Portion of CD46 Provides Insights into Its Interactions with Complement Proteins and Pathogens. PLoS Pathog. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. - DOI - PMC - PubMed
-
- Post T.W., Liszewski M.K., Adams E.M., Tedja I., Miller E.A., Atkinson J.P. Membrane cofactor protein of the complement system: Alternative splicing of serine/threonine/proline-rich exons and cytoplasmic tails produces multiple isoforms that correlate with protein phenotype. J. Exp. Med. 1991;174:93–102. doi: 10.1084/jem.174.1.93. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

