Cytotoxic T Lymphocytes Control Growth of B16 Tumor Cells in Collagen-Fibrin Gels by Cytolytic and Non-Lytic Mechanisms
- PMID: 37515143
- PMCID: PMC10384826
- DOI: 10.3390/v15071454
Cytotoxic T Lymphocytes Control Growth of B16 Tumor Cells in Collagen-Fibrin Gels by Cytolytic and Non-Lytic Mechanisms
Abstract
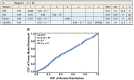
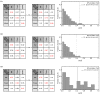
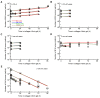
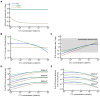
Cytotoxic T lymphocytes (CTLs) are important in controlling some viral infections, and therapies involving the transfer of large numbers of cancer-specific CTLs have been successfully used to treat several types of cancers in humans. While the molecular mechanisms of how CTLs kill their targets are relatively well understood, we still lack a solid quantitative understanding of the kinetics and efficiency by which CTLs kill their targets in vivo. Collagen-fibrin-gel-based assays provide a tissue-like environment for the migration of CTLs, making them an attractive system to study T cell cytotoxicity in in vivo-like conditions. Budhu.et al. systematically varied the number of peptide (SIINFEKL)-pulsed B16 melanoma cells and SIINFEKL-specific CTLs (OT-1) and measured the remaining targets at different times after target and CTL co-inoculation into collagen-fibrin gels. The authors proposed that their data were consistent with a simple model in which tumors grow exponentially and are killed by CTLs at a per capita rate proportional to the CTL density in the gel. By fitting several alternative mathematical models to these data, we found that this simple "exponential-growth-mass-action-killing" model did not precisely describe the data. However, determining the best-fit model proved difficult because the best-performing model was dependent on the specific dataset chosen for the analysis. When considering all data that include biologically realistic CTL concentrations (E≤107cell/mL), the model in which tumors grow exponentially and CTLs suppress tumor's growth non-lytically and kill tumors according to the mass-action law (SiGMA model) fit the data with the best quality. A novel power analysis suggested that longer experiments (∼3-4 days) with four measurements of B16 tumor cell concentrations for a range of CTL concentrations would best allow discriminating between alternative models. Taken together, our results suggested that the interactions between tumors and CTLs in collagen-fibrin gels are more complex than a simple exponential-growth-mass-action killing model and provide support for the hypothesis that CTLs' impact on tumors may go beyond direct cytotoxicity.
Keywords: B16 tumors; cytotoxic T lymphocytes; killing; mathematical modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Update of
-
Mathematical modeling suggests cytotoxic T lymphocytes control growth of B16 tumor cells in collagin-fibrin gels by cytolytic and non-lytic mechanisms.bioRxiv [Preprint]. 2023 Mar 29:2023.03.28.534600. doi: 10.1101/2023.03.28.534600. bioRxiv. 2023. Update in: Viruses. 2023 Jun 27;15(7):1454. doi: 10.3390/v15071454. PMID: 37034693 Free PMC article. Updated. Preprint.
References
-
- Halle S., Keyser K.A., Stahl F.R., Busche A., Marquardt A., Zheng X., Galla M., Heissmeyer V., Heller K., Boelter J., et al. In Vivo Killing Capacity of Cytotoxic T Cells Is Limited and Involves Dynamic Interactions and T Cell Cooperativity. Immunity. 2016;44:233–245. doi: 10.1016/j.immuni.2016.01.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

