Lumpy Skin Disease Virus Genome Sequence Analysis: Putative Spatio-Temporal Epidemiology, Single Gene versus Whole Genome Phylogeny and Genomic Evolution
- PMID: 37515159
- PMCID: PMC10385495
- DOI: 10.3390/v15071471
Lumpy Skin Disease Virus Genome Sequence Analysis: Putative Spatio-Temporal Epidemiology, Single Gene versus Whole Genome Phylogeny and Genomic Evolution
Abstract
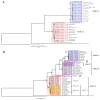
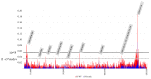
Lumpy Skin Disease virus is a poxvirus from the genus Capripox that mainly affects bovines and it causes severe economic losses to livestock holders. The Lumpy Skin Disease virus is currently dispersing in Asia, but little is known about detailed phylogenetic relations between the strains and genome evolution. We reconstructed a whole-genome-sequence (WGS)-based phylogeny and compared it with single-gene-based phylogenies. To study population and spatiotemporal patterns in greater detail, we reconstructed networks. We determined that there are strains from multiple clades within the previously defined cluster 1.2 that correspond with recorded outbreaks across Eurasia and South Asia (Indian subcontinent), while strains from cluster 2.5 spread in Southeast Asia. We concluded that using only a single gene (cheap, fast and easy to routinely use) for sequencing lacks phylogenetic and spatiotemporal resolution and we recommend to create at least one WGS whenever possible. We also found that there are three gene regions, highly variable, across the genome of LSDV. These gene regions are located in the 5' and 3' flanking regions of the LSDV genome and they encode genes that are involved in immune evasion strategies of the virus. These may provide a starting point to further investigate the evolution of the virus.
Keywords: evolution; genome; lumpy skin disease; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Sohier C., Haegeman A., Mostin L., De Leeuw I., Campe W.V., De Vleeschauwer A., Tuppurainen E.S.M., van den Berg T., De Regge N., De Clercq K. Experimental evidence of mechanical Lumpy Skin Disease virus transmission by Stomoxys calcitrans biting flies and Haematopota spp. horseflies. Sci. Rep. 2019;9:20076. doi: 10.1038/s41598-019-56605-6. - DOI - PMC - PubMed
-
- Coetzer J.A.W., Tuppurainen E., Babiuk S., Wallace D. Infectious Diseases of Livestock, Part II. Anipedia; Pretoria, South Africa: 2018. Lumpy Skin Disease.
-
- Kononov A., Byadovskaya O., Wallace D., Prutnikov P., Pestova Y., Kononova S., Nesterov A., Rusaleev V., Lozovoy D., Sprygin A. Non-vector-borne transmission of Lumpy Skin Disease virus. Sci. Rep. 2020;10:7436.
MeSH terms
LinkOut - more resources
Full Text Sources

