Diphtheria toxin-derived, anti-PD-1 immunotoxin, a potent and practical tool to selectively deplete PD-1+ cells
- PMID: 37515422
- PMCID: PMC10443333
- DOI: 10.1002/pro.4741
Diphtheria toxin-derived, anti-PD-1 immunotoxin, a potent and practical tool to selectively deplete PD-1+ cells
Abstract
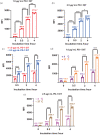
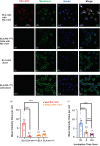
Programmed death-1 (PD-1), an immune checkpoint receptor, is expressed on activated lymphocytes, macrophages, and some types of tumor cells. While PD-1+ cells have been implicated in outcomes of cancer immunity, autoimmunity, and chronic infections, the exact roles of these cells in various physiological and pathological processes remain elusive. Molecules that target and deplete PD-1+ cells would be instrumental in defining the roles unambiguously. Previously, an immunotoxin has been generated for the depletion of PD-1+ cells though its usage is impeded by its low production yield. Thus, a more practical molecular tool is desired to deplete PD-1+ cells and to examine functions of these cells. We designed and generated a novel anti-PD1 diphtheria immunotoxin, termed PD-1 DIT, targeting PD-1+ cells. PD-1 DIT is comprised of two single chain variable fragments (scFv) derived from an anti-PD-1 antibody, coupled with the catalytic and translocation domains of the diphtheria toxin. PD-1 DIT was produced using a yeast expression system that has been engineered to efficiently produce protein toxins. The yield of PD-1 DIT reached 1-2 mg/L culture, which is 10 times higher than the previously reported immunotoxin. Flow cytometry and confocal microscopy analyses confirmed that PD-1 DIT specifically binds to and enters PD-1+ cells. The binding avidities between PD-1 DIT and two PD-1+ cell lines are approximately 25 nM. Moreover, PD-1 DIT demonstrated potent cytotoxicity toward PD-1+ cells, with a half maximal effective concentration (EC50 ) value of 1 nM. In vivo experiments further showed that PD-1 DIT effectively depleted PD-1+ cells and enabled mice inoculated with PD-1+ tumor cells to survive throughout the study. Our findings using PD-1 DIT revealed the critical role of pancreatic PD-1+ T cells in the development of type-1 diabetes (T1D). Additionally, we observed that PD-1 DIT treatment ameliorated relapsing-remitting experimental autoimmune encephalomyelitis (RR-EAE), a mouse model of relapsing-remitting multiple sclerosis (RR-MS). Lastly, we did not observe significant hepatotoxicity in mice treated with PD-1 DIT, which had been reported for other immunotoxins derived from the diphtheria toxin. With its remarkable selective and potent cytotoxicity toward PD-1+ cells, coupled with its high production yield, PD-1 DIT emerges as a powerful biotechnological tool for elucidating the physiological roles of PD-1+ cells. Furthermore, the potential of PD-1 DIT to be developed into a novel therapeutic agent becomes evident.
Keywords: PD-1; autoimmune diseases; cancer; diphtheria toxin-derived immunotoxin; programmed death-1 cells; protein therapeutics; yeast.
© 2023 The Protein Society.
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures




References
-
- Arata Y, Watanabe A, Motosugi R, Murakami R, Goto T, Hori S, et al. Defective induction of the proteasome associated with T‐cell receptor signaling underlies T‐cell senescence. Genes Cells. 2019;24(12):801–813. - PubMed
-
- Arndt Br, Witkowski L, Ellwart J, Seissler J. CD8+ CD122+ PD‐1− effector cells promote the development of diabetes in NOD mice. J Leucocyte Biol. 2015;97(1):111–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

