Cryo-EM structure of a RAS/RAF recruitment complex
- PMID: 37516774
- PMCID: PMC10387098
- DOI: 10.1038/s41467-023-40299-6
Cryo-EM structure of a RAS/RAF recruitment complex
Erratum in
-
Publisher Correction: Cryo-EM structure of a RAS/RAF recruitment complex.Nat Commun. 2023 Sep 7;14(1):5517. doi: 10.1038/s41467-023-41365-9. Nat Commun. 2023. PMID: 37679375 Free PMC article. No abstract available.
Abstract
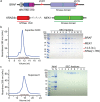
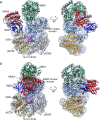
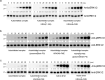
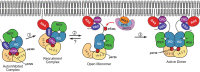
RAF-family kinases are activated by recruitment to the plasma membrane by GTP-bound RAS, whereupon they initiate signaling through the MAP kinase cascade. Prior structural studies of KRAS with RAF have focused on the isolated RAS-binding and cysteine-rich domains of RAF (RBD and CRD, respectively), which interact directly with RAS. Here we describe cryo-EM structures of a KRAS bound to intact BRAF in an autoinhibited state with MEK1 and a 14-3-3 dimer. Analysis of this KRAS/BRAF/MEK1/14-3-3 complex reveals KRAS bound to the RAS-binding domain of BRAF, captured in two orientations. Core autoinhibitory interactions in the complex are unperturbed by binding of KRAS and in vitro activation studies confirm that KRAS binding is insufficient to activate BRAF, absent membrane recruitment. These structures illustrate the separability of binding and activation of BRAF by RAS and suggest stabilization of this pre-activation intermediate as an alternative therapeutic strategy to blocking binding of KRAS.
© 2023. The Author(s).
Conflict of interest statement
M.J.E. receives or has received sponsored research support from Novartis, Sanofi, Takeda, and Springworks Therapeutics and consulting income or honoraria from Novartis, H3 Biomedicine and Ikena Oncology. The remaining authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

