METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer
- PMID: 37516825
- PMCID: PMC10386714
- DOI: 10.1186/s12943-023-01809-8
METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer
Abstract
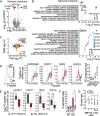
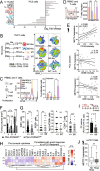
Newly growing evidence highlights the essential role that epitranscriptomic marks play in the development of many cancers; however, little is known about the role and implications of altered epitranscriptome deposition in prostate cancer. Here, we show that the transfer RNA N7-methylguanosine (m7G) transferase METTL1 is highly expressed in primary and advanced prostate tumours. Mechanistically, we find that METTL1 depletion causes the loss of m7G tRNA methylation and promotes the biogenesis of a novel class of small non-coding RNAs derived from 5'tRNA fragments. 5'tRNA-derived small RNAs steer translation control to favour the synthesis of key regulators of tumour growth suppression, interferon pathway, and immune effectors. Knockdown of Mettl1 in prostate cancer preclinical models increases intratumoural infiltration of pro-inflammatory immune cells and enhances responses to immunotherapy. Collectively, our findings reveal a therapeutically actionable role of METTL1-directed m7G tRNA methylation in cancer cell translation control and tumour biology.
Keywords: 7-methylguanosine; Epitranscriptome; Immune checkpoint blockade; Interferon; Prostate cancer; RNA modifications; Tumour microenvironment (TME); tRNA fragments.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387:70–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

