HSV-1 exploits host heterochromatin for nuclear egress
- PMID: 37516914
- PMCID: PMC10373338
- DOI: 10.1083/jcb.202304106
HSV-1 exploits host heterochromatin for nuclear egress
Abstract
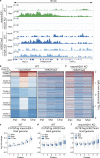
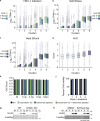
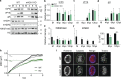
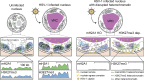
Herpes simplex virus (HSV-1) progeny form in the nucleus and exit to successfully infect other cells. Newly formed capsids navigate complex chromatin architecture to reach the inner nuclear membrane (INM) and egress. Here, we demonstrate by transmission electron microscopy (TEM) that HSV-1 capsids traverse heterochromatin associated with trimethylation on histone H3 lysine 27 (H3K27me3) and the histone variant macroH2A1. Through chromatin profiling during infection, we revealed global redistribution of these marks whereby massive host genomic regions bound by macroH2A1 and H3K27me3 correlate with decreased host transcription in active compartments. We found that the loss of these markers resulted in significantly lower viral titers but did not impact viral genome or protein accumulation. Strikingly, we discovered that loss of macroH2A1 or H3K27me3 resulted in nuclear trapping of capsids. Finally, by live-capsid tracking, we quantified this decreased capsid movement. Thus, our work demonstrates that HSV-1 takes advantage of the dynamic nature of host heterochromatin formation during infection for efficient nuclear egress.
© 2023 Lewis et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







References
-
- Aho, V., Mäntylä E., Ekman A., Hakanen S., Mattola S., Chen J.-H., Weinhardt V., Ruokolainen V., Sodeik B., Larabell C., and Vihinen-Ranta M.. 2019. Quantitative microscopy reveals stepwise alteration of chromatin structure during herpesvirus infection. Viruses. 11:935. 10.3390/v11100935 - DOI - PMC - PubMed
-
- Aho, V., Salminen S., Mattola S., Gupta A., Flomm F., Sodeik B., Bosse J.B., and Vihinen-Ranta M.. 2021. Infection-induced chromatin modifications facilitate translocation of herpes simplex virus capsids to the inner nuclear membrane. PLoS Pathog. 17:e1010132. 10.1371/journal.ppat.1010132 - DOI - PMC - PubMed
-
- Arbuckle, J.H., Gardina P.J., Gordon D.N., Hickman H.D., Yewdell J.W., Pierson T.C., Myers T.G., and Kristie T.M.. 2017. Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. MBio. 8:e01141-17. 10.1128/mBio.01141-17 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

