Development of a V5-tag-directed nanobody and its implementation as an intracellular biosensor of GPCR signaling
- PMID: 37517699
- PMCID: PMC10470007
- DOI: 10.1016/j.jbc.2023.105107
Development of a V5-tag-directed nanobody and its implementation as an intracellular biosensor of GPCR signaling
Abstract
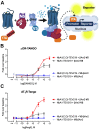
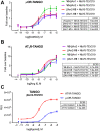
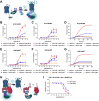
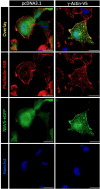
Protein-protein interactions (PPIs) form the foundation of any cell signaling network. Considering that PPIs are highly dynamic processes, cellular assays are often essential for their study because they closely mimic the biological complexities of cellular environments. However, incongruity may be observed across different PPI assays when investigating a protein partner of interest; these discrepancies can be partially attributed to the fusion of different large functional moieties, such as fluorescent proteins or enzymes, which can yield disparate perturbations to the protein's stability, subcellular localization, and interaction partners depending on the given cellular assay. Owing to their smaller size, epitope tags may exhibit a diminished susceptibility to instigate such perturbations. However, while they have been widely used for detecting or manipulating proteins in vitro, epitope tags lack the in vivo traceability and functionality needed for intracellular biosensors. Herein, we develop NbV5, an intracellular nanobody binding the V5-tag, which is suitable for use in cellular assays commonly used to study PPIs such as BRET, NanoBiT, and Tango. The NbV5:V5 tag system has been applied to interrogate G protein-coupled receptor signaling, specifically by replacing larger functional moieties attached to the protein interactors, such as fluorescent or luminescent proteins (∼30 kDa), by the significantly smaller V5-tag peptide (1.4 kDa), and for microscopy imaging which is successfully detected by NbV5-based biosensors. Therefore, the NbV5:V5 tag system presents itself as a versatile tool for live-cell imaging and a befitting adaptation to existing cellular assays dedicated to probing PPIs.
Keywords: G protein-coupled receptor (GPCR); bioluminescence resonance energy transfer (BRET); crystal structure; nanobody); nanoluciferase binary technology (NanoBiT); protease-dependent reporter assay (Tango); protein-protein interaction; single-domain antibody (sdAb.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests The authors declare that they have no conflicts of interests with the contents of this article.
Figures



References
-
- Milligan G., andWhite J.H. Protein-protein interactions at G-protein-coupled receptors. Trends Pharmacol. Sci. 2001;22:513–518. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

