Epigenetics in myeloproliferative neoplasms
- PMID: 37519812
- PMCID: PMC10373880
- DOI: 10.3389/fonc.2023.1206965
Epigenetics in myeloproliferative neoplasms
Abstract
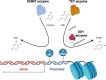
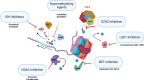
The myeloproliferative neoplasms (MPNs) are a group of acquired clonal disorders where mutations drive proliferative disease resulting in increased blood counts and in some cases end-stage myelofibrosis. Epigenetic changes are the reversible modifications to DNA- and RNA-associated proteins that impact gene activity without changing the DNA sequence. This review summarizes mechanisms of epigenetic changes and the nucleosome. The drivers and epigenetic regulators in MPNs are outlined. In MPNs, distinct patterns of epigenetic dysregulation have been seen in chronic and in advanced phases. Methylation age and histone modification are altered in MPNs and by further treatment. The alterations found in methylation age in MPNs and with treatment are discussed, and the changes in histone modification with Janus kinase (JAK) inhibition are evaluated. Currently available therapeutic areas where the epigenome can be altered are outlined. Thus, we review the current knowledge and understanding of epigenetics in MPN and consider further management options. Understanding the epigenome and its alteration in MPNs and epigenetic changes associated with the progression of disease will lead to advances in therapeutic options.
Keywords: DNA methylation; JAK2; NFE2; epigenetics; histone modification; myeloproliferative neoplasms.
Copyright © 2023 Greenfield and McMullin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
-
Unfolding the pathogenesis of scleroderma through genomics and epigenomics.J Autoimmun. 2017 Sep;83:73-94. doi: 10.1016/j.jaut.2017.05.004. Epub 2017 May 16. J Autoimmun. 2017. PMID: 28526340 Free PMC article.
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
Epigenetic abnormalities in myeloproliferative neoplasms: a target for novel therapeutic strategies.Clin Epigenetics. 2011 Aug;2(2):197-212. doi: 10.1007/s13148-011-0050-6. Epub 2011 Jul 9. Clin Epigenetics. 2011. PMID: 22704337 Free PMC article.
Cited by
-
Sphingosine kinase 1 facilitates gastric cancer progression via STAT1 gene methylation-mediated mechanisms.Transl Cancer Res. 2025 May 30;14(5):3226-3238. doi: 10.21037/tcr-2025-948. Epub 2025 May 27. Transl Cancer Res. 2025. PMID: 40530131 Free PMC article.
-
DNA Methylation in Ph-Negative Myeloproliferative Neoplasms: Prognostic Role and Therapeutic Implications.Curr Issues Mol Biol. 2025 Mar 26;47(4):227. doi: 10.3390/cimb47040227. Curr Issues Mol Biol. 2025. PMID: 40699626 Free PMC article. Review.
-
Single nucleotide polymorphism and promoter methylation analysis of protein tyrosine phosphatase 1B in patients with myeloproliferative neoplasms.Transl Cancer Res. 2025 Jan 31;14(1):212-224. doi: 10.21037/tcr-24-1338. Epub 2025 Jan 23. Transl Cancer Res. 2025. PMID: 39974415 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

