sgRNAs: A SARS-CoV-2 emerging issue
- PMID: 37519862
- PMCID: PMC10105645
- DOI: 10.1016/j.amolm.2023.100008
sgRNAs: A SARS-CoV-2 emerging issue
Abstract
Like for other coronaviruses, SARS-CoV-2 gene expression strategy is based on the synthesis of a nested set of subgenomic mRNA species (sgRNAs). These sgRNA are synthesized using a "discontinuous transcription" mechanism that relies on template switching at Transcription Regulatory Sequences (TRS). Both canonical (c-sgRNA) and non-canonical (nc-sgRNA, less numerous) subgenomic RNA species can be produced. Currently, sgRNAs are investigated on the basis of sequence data obtained through next generation sequencing (NGS), and bioinformatic tools are crucial for their identification, characterization and quantification. To date, few software have been developed to this aim, whose reliability and applicability to all the available NGS platforms need to be established, to build confidence on the information resulting from such tools. In fact, these information may be crucial for the in depth elucidation of viral expression strategy, particularly in respect of the significance of nc-sgRNAs, and for the possible use of sgRNAs as potential markers of virus replicative activity in infected patients.
Keywords: Bioinformatics; Coronavirus; Next-generation sequencing; SARS-CoV2; Viruses; sgRNAs.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
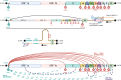
Figures
References
-
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Hüttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Félix E., Shi Y., Goff M., Lim J.K., McBride T., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., De wit E., Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B.R., Mullins R.D., Fischer E.R., Kochs G., Grosse R., García-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/J.CELL.2020.06.034/ATTACHMENT/2523A3EE-6406-4D91-B592-8E7E65AB0F71/MMC8.XLSX. - DOI - PMC - PubMed
-
- Chang J.J.Y., Rawlinson D., Pitt M.E., Taiaroa G., Gleeson J., Zhou C., Mordant F.L., De Paoli-Iseppi R., Caly L., Purcell D.F.J., Stinear T.P., Londrigan S.L., Clark M.B., Williamson D.A., Subbarao K., Coin L.J.M. Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection. Cell Rep. 2021;35 doi: 10.1016/J.CELREP.2021.109108. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
