Screening, molecular identification, population diversity, and antibiotic susceptibility pattern of Actinomycetes species isolated from meat and meat products of slaughterhouses, restaurants, and meat stores of a developing country, Iran
- PMID: 37520382
- PMCID: PMC10373891
- DOI: 10.3389/fmicb.2023.1134368
Screening, molecular identification, population diversity, and antibiotic susceptibility pattern of Actinomycetes species isolated from meat and meat products of slaughterhouses, restaurants, and meat stores of a developing country, Iran
Abstract
Introduction: Actinomycetes can colonize surfaces of tools and equipment and can be transferred to meat and meat products during manufacture, processing, handling, and storage. Moreover, washing the meat does not eliminate the microorganisms; it only spreads them. As a result, these opportunistic pathogens can enter the human body and cause various infections. Therefore, the aim of the current study was to screen, identify, and determine the antibiotic susceptibility of Actinomycetes species from meat and meat products in the Markazi province of Iran.
Methods: A total of 60 meat and meat product samples, including minced meat, mutton, beef, chicken, hamburgers, and sausages, were collected from slaughterhouses, butchers, and restaurants in the Markazi province of Iran. The samples were analyzed using standard microbiological protocols for the isolation and characterization of Actinomycetes. PCR amplification of hsp65 and 16SrRNA genes and sequence analysis of 16SrRNA were used for genus and species identification. The minimum inhibitory concentrations (MICs) of antimicrobial agents were determined by the broth microdilution method and interpreted according to the CLSI guidelines.
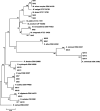
Results: A total of 21 (35%) Actinomycetes isolates from 5 genera and 12 species were isolated from 60 samples. The most prevalent Actinomycetes were from the genus Mycobacterium, with six (28.6%) isolates (M. avium complex, M. terrae, M. smegmatis, and M. novocastrense), followed by the genus Rhodococcus with five (23.8%) isolates (R. equi and R. erythropolis), the genus Actinomyces with four (19.1%) isolates (A. ruminicola and A. viscosus), the genus Nocardia with four (19.1%) isolates (N. asiatica, N. seriolae, and N. niigatensis), and the genus Streptomyces with two (9.5%) isolates (S. albus). Chicken and sausage samples had the highest and lowest levels of contamination, with six and one isolates. Respectively, the results of drug susceptibility testing (DST) showed that all isolates were susceptible to Ofloxacin, Amikacin, Ciprofloxacin, and Levofloxacin, whereas all of them were resistant to Doxycycline and Rifampicin.
Discussion: The findings suggest that meat and meat products play an important role as a reservoir for the transmission of Actinomycetes to humans, thus causing life-threatening foodborne diseases such as gastrointestinal and cutaneous disorders. Therefore, it is essential to incorporate basic hygiene measures into the cycle of meat production to ensure food safety.
Keywords: 16SrRNA; Actinomycetes; DST; food born infection; meat and meat product.
Copyright © 2023 Motallebirad, Mardanshah, Safarabadi, Ghaffari, Orouji, Abedi and Azadi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Species Diversity, Molecular Characterization, and Antimicrobial Susceptibility of Opportunistic Actinomycetes Isolated from Immunocompromised and Healthy Patients of Markazi Province of Iran.Infect Drug Resist. 2020 Jan 7;13:1-10. doi: 10.2147/IDR.S234292. eCollection 2020. Infect Drug Resist. 2020. PMID: 32021315 Free PMC article.
-
Species diversity and molecular analysis of opportunistic Mycobacterium, Nocardia and Rhodococcus isolated from the hospital environment in a developing country, a potential resources for nosocomial infection.Genes Environ. 2021 Jan 28;43(1):2. doi: 10.1186/s41021-021-00173-7. Genes Environ. 2021. PMID: 33509299 Free PMC article.
-
Molecular identification and antibiotic resistance pattern of actinomycetes isolates among immunocompromised patients in Iran, emerging of new infections.Sci Rep. 2021 May 24;11(1):10745. doi: 10.1038/s41598-021-90269-5. Sci Rep. 2021. PMID: 34031507 Free PMC article.
-
Fifteen years of phenotypic and genotypic surveillance and antibiotic susceptibility pattern of Actinomycetes (Mycobacterium, Nocardia, Rhodococcus, etc.) in clinical and environmental samples of Iran.Diagn Microbiol Infect Dis. 2024 Jan;108(1):116080. doi: 10.1016/j.diagmicrobio.2023.116080. Epub 2023 Sep 9. Diagn Microbiol Infect Dis. 2024. PMID: 37862765 Review.
-
Candida and candidaemia. Susceptibility and epidemiology.Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
Cited by
-
Research on Bacterial Diversity and Antibiotic Resistance in the Dairy Farm Environment in a Part of Shandong Province.Animals (Basel). 2024 Jan 3;14(1):160. doi: 10.3390/ani14010160. Animals (Basel). 2024. PMID: 38200891 Free PMC article.
-
Tracheal tube infections in critical care: A narrative review of influencing factors, microbial agents, and mitigation strategies in intensive care unit settings.SAGE Open Med. 2024 Dec 16;12:20503121241306951. doi: 10.1177/20503121241306951. eCollection 2024. SAGE Open Med. 2024. PMID: 39691863 Free PMC article. Review.
-
Exploring the Potential of Halotolerant Actinomycetes from Rann of Kutch, India: A Study on the Synthesis, Characterization, and Biomedical Applications of Silver Nanoparticles.Pharmaceuticals (Basel). 2024 Jun 6;17(6):743. doi: 10.3390/ph17060743. Pharmaceuticals (Basel). 2024. PMID: 38931410 Free PMC article.
-
Molecular surveillance and antimicrobial susceptibility profile of bacterial contamination in pastries of Iranian confectioneries: a public health concern.Front Microbiol. 2024 Dec 4;15:1494623. doi: 10.3389/fmicb.2024.1494623. eCollection 2024. Front Microbiol. 2024. PMID: 39697660 Free PMC article.
References
-
- Anandan R., Dharumadurai D., Manogaran G. P. (2016). “An introduction to actinobacteria” in Actinobacteria-basics and biotechnological applications. eds. Dhanasekaran D., Jiang Y. (London: IntechOpen; )
-
- Azadi D., Motallebirad T., Ghaffari K., Shokri D., Rezaei F. (2020). Species diversity, molecular characterization, and antimicrobial susceptibility of opportunistic Actinomycetes isolated from immunocompromised and healthy patients of Markazi Province of Iran. Infect. Drug Resist. 13, 1–10. doi: 10.2147/IDR.S234292, PMID: - DOI - PMC - PubMed
-
- Azadi D., Shojaei H., Pourchangiz M., Dibaj R., Davarpanah M., Naser A. D. (2016). Species diversity and molecular characterization of nontuberculous mycobacteria in hospital water system of a developing country, Iran. Microb. Pathog. 100, 62–69. doi: 10.1016/j.micpath.2016.09.004, PMID: - DOI - PubMed
-
- Bakhtiary F., Sayevand H. R., Remely M., Hippe B., Hosseini H., Haslberger A. G. (2016). Evaluation of bacterial contamination sources in meat production line. J. Food Qual. 39, 750–756. doi: 10.1111/jfq.12243 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials