circIFNGR2 regulating ankylosing spondylitis-associated inflammation through macrophage polarization
- PMID: 37520722
- PMCID: PMC10372825
- DOI: 10.1016/j.isci.2023.107325
circIFNGR2 regulating ankylosing spondylitis-associated inflammation through macrophage polarization
Abstract
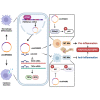
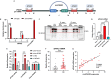
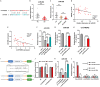
Macrophages activation is crucial in pathogenesis of rheumatic diseases like ankylosing spondylitis (AS). Circular RNAs (circRNAs)-induced macrophage-associated inflammation participates in many autoimmune diseases but remains elusive in AS. Here, we verified increased expression of circIFNGR2 in peripheral blood mononuclear cells from patients with AS and its expression levels were correlated with the AS severity. In vitro assays revealed that circIFNGR2 enhances macrophage proliferation, and regulates M1/M2 macrophage polarization and NF-κB/Akt pathways. We identified that circIFNGR2 promoted the expression of iNOS/TNFα and M1 polarization, and restrained M2 polarization by sponging miR-939. Additionally, the RNA-binding protein, eIF4A3, was found to enhance the production of circIFNGR2. Interestingly, miR-939 attenuated joint damage in collagen-induced arthritis mice, whereas circIFNGR2 reversed this effect. Our findings highlight the pro-inflammatory roles of eIF4A3-induced circIFNGR2 in AS by modulating macrophage-associated inflammation through miR-939.
Keywords: Clinical genetics; Disease; Pathophysiology.
© 2023 The Authors.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

