Connectome-based machine learning models are vulnerable to subtle data manipulations
- PMID: 37521052
- PMCID: PMC10382940
- DOI: 10.1016/j.patter.2023.100756
Connectome-based machine learning models are vulnerable to subtle data manipulations
Abstract
Neuroimaging-based predictive models continue to improve in performance, yet a widely overlooked aspect of these models is "trustworthiness," or robustness to data manipulations. High trustworthiness is imperative for researchers to have confidence in their findings and interpretations. In this work, we used functional connectomes to explore how minor data manipulations influence machine learning predictions. These manipulations included a method to falsely enhance prediction performance and adversarial noise attacks designed to degrade performance. Although these data manipulations drastically changed model performance, the original and manipulated data were extremely similar (r = 0.99) and did not affect other downstream analysis. Essentially, connectome data could be inconspicuously modified to achieve any desired prediction performance. Overall, our enhancement attacks and evaluation of existing adversarial noise attacks in connectome-based models highlight the need for counter-measures that improve the trustworthiness to preserve the integrity of academic research and any potential translational applications.
Keywords: adversarial attacks; connectomics; fMRI; functional connectivity; machine learning; predictive modeling; trustworthiness.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





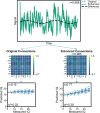
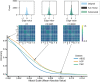
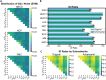
References
Grants and funding
- U24 DA041147/DA/NIDA NIH HHS/United States
- U01 DA041120/DA/NIDA NIH HHS/United States
- T32 DA022975/DA/NIDA NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U24 DA041123/DA/NIDA NIH HHS/United States
- RC2 MH089924/MH/NIMH NIH HHS/United States
- U01 DA041156/DA/NIDA NIH HHS/United States
- U01 DA041106/DA/NIDA NIH HHS/United States
- U01 DA041117/DA/NIDA NIH HHS/United States
- U01 DA041148/DA/NIDA NIH HHS/United States
- K00 MH122372/MH/NIMH NIH HHS/United States
- RC2 MH089983/MH/NIMH NIH HHS/United States
- U01 DA041174/DA/NIDA NIH HHS/United States
- U01 DA041134/DA/NIDA NIH HHS/United States
- U01 DA041022/DA/NIDA NIH HHS/United States
- T32 GM100884/GM/NIGMS NIH HHS/United States
- U54 MH091657/MH/NIMH NIH HHS/United States
- U01 DA041089/DA/NIDA NIH HHS/United States
- R01 MH121095/MH/NIMH NIH HHS/United States
- U01 DA041028/DA/NIDA NIH HHS/United States
- U01 DA041048/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources

