From endodormancy to ecodormancy: the transcriptional landscape of apple floral buds
- PMID: 37521930
- PMCID: PMC10375413
- DOI: 10.3389/fpls.2023.1194244
From endodormancy to ecodormancy: the transcriptional landscape of apple floral buds
Abstract
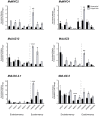
This study endeavors to explore the transcriptomic profiles of two apple cultivars, namely, 'Honeycrisp' and 'Cripps Pink,' which represent late and early-blooming cultivars, respectively. Using RNA-sequencing technology, we analyzed floral bud samples collected at five distinct time intervals during both endodormancy and ecodormancy. To evaluate the transcriptomic profiles of the 30 sequenced samples, we conducted principal component analysis (PCA). PC1 explained 43% of the variance, separating endodormancy and ecodormancy periods, while PC2 explained 16% of the variance, separating the two cultivars. The number of differentially expressed genes (DEGs) increased with endodormancy progression and remained elevated during ecodormancy. The majority of DEGs were unique to a particular time point, with only a few overlapping among or between the time points. This highlights the temporal specificity of gene expression during the dormancy transition and emphasizes the importance of sampling at multiple time points to capture the complete transcriptomic dynamics of this intricate process. We identified a total of 4204 upregulated and 7817 downregulated DEGs in the comparison of endodormancy and ecodormancy, regardless of cultivar, and 2135 upregulated and 2413 downregulated DEGs in the comparison of 'Honeycrisp' versus 'Cripps Pink,' regardless of dormancy stage. Furthermore, we conducted a co-expression network analysis to gain insight into the coordinated gene expression profiles across different time points, dormancy stages, and cultivars. This analysis revealed the most significant module (ME 14), correlated with 1000 GDH and consisting of 1162 genes. The expression of the genes within this module was lower in 'Honeycrisp' than in 'Cripps Pink.' The top 20 DEGs identified in ME 14 were primarily related to jasmonic acid biosynthesis and signaling, lipid metabolism, oxidation-reduction, and transmembrane transport activity. This suggests a plausible role for these pathways in governing bud dormancy and flowering time in apple.
Keywords: bud dormancy; ecodormancy; endodormancy; flowering time; jasmonic acid; transcriptomics.
Copyright © 2023 Sapkota, Salem, Jahed, Artlip and Sherif.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
-
- Bielenberg D. G., Wang Y., Li Z., Zhebentyayeva T., Fan S., Reighard G. L., et al. . (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507. doi: 10.1007/s11295-007-0126-9 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

