Human herpesvirus 8 ORF57 protein is able to reduce TDP-43 pathology: network analysis identifies interacting pathways
- PMID: 37522762
- PMCID: PMC10549787
- DOI: 10.1093/hmg/ddad122
Human herpesvirus 8 ORF57 protein is able to reduce TDP-43 pathology: network analysis identifies interacting pathways
Abstract
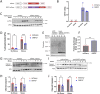
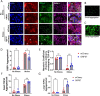
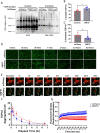
Aggregation of TAR DNA-binding protein 43 kDa (TDP-43) is thought to drive the pathophysiology of amyotrophic lateral sclerosis and some frontotemporal dementias. TDP-43 is normally a nuclear protein that in neurons translocates to the cytoplasm and can form insoluble aggregates upon activation of the integrated stress response (ISR). Viruses evolved to control the ISR. In the case of Herpesvirus 8, the protein ORF57 acts to bind protein kinase R, inhibit phosphorylation of eIF2α and reduce activation of the ISR. We hypothesized that ORF57 might also possess the ability to inhibit aggregation of TDP-43. ORF57 was expressed in the neuronal SH-SY5Y line and its effects on TDP-43 aggregation characterized. We report that ORF57 inhibits TDP-43 aggregation by 55% and elicits a 2.45-fold increase in the rate of dispersion of existing TDP-43 granules. These changes were associated with a 50% decrease in cell death. Proteomic studies were carried out to identify the protein interaction network of ORF57. We observed that ORF57 directly binds to TDP-43 as well as interacts with many components of the ISR, including elements of the proteostasis machinery known to reduce TDP-43 aggregation. We propose that viral proteins designed to inhibit a chronic ISR can be engineered to remove aggregated proteins and dampen a chronic ISR.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Neumann, M., Sampathu, D.M., Kwong, L.K., Truax, A.C., Micsenyi, M.C., Chou, T.T., Bruce, J., Schuck, T., Grossman, M., Clark, C.M. et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science, 314, 130–133. - PubMed
-
- Igaz, L.M., Kwong, L.K., Xu, Y., Truax, A.C., Uryu, K., Neumann, M., Clark, C.M., Elman, L.B., Miller, B.L., Grossman, M. et al. (2008) Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol., 173, 182–194. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

