Embryo-uterine interaction coordinates mouse embryogenesis during implantation
- PMID: 37522872
- PMCID: PMC10476174
- DOI: 10.15252/embj.2022113280
Embryo-uterine interaction coordinates mouse embryogenesis during implantation
Abstract
Embryo implantation into the uterus marks a key transition in mammalian development. In mice, implantation is mediated by the trophoblast and is accompanied by a morphological transition from the blastocyst to the egg cylinder. However, the roles of trophoblast-uterine interactions in embryo morphogenesis during implantation are poorly understood due to inaccessibility in utero and the remaining challenges to recapitulate it ex vivo from the blastocyst. Here, we engineer a uterus-like microenvironment to recapitulate peri-implantation development of the whole mouse embryo ex vivo and reveal essential roles of the physical embryo-uterine interaction. We demonstrate that adhesion between the trophoblast and the uterine matrix is required for in utero-like transition of the blastocyst to the egg cylinder. Modeling the implanting embryo as a wetting droplet links embryo shape dynamics to the underlying changes in trophoblast adhesion and suggests that the adhesion-mediated tension release facilitates egg cylinder formation. Light-sheet live imaging and the experimental control of the engineered uterine geometry and trophoblast velocity uncovers the coordination between trophoblast motility and embryo growth, where the trophoblast delineates space for embryo morphogenesis.
Keywords: Implantation; biophysical modeling; embryo; engineering; uterus.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors filed a patent EP23162464.4 devoted to the live imaging and embryo culture method.
Figures

- A
Schematic showing the lineages and the layers of extracellular matrix, comprising in utero peri‐implantation mouse embryo at E5.25. EPI, epiblast; ExE, extraembryonic ectoderm; TB, trophoblast; PE, parietal endoderm; VE, visceral endoderm; BM, the basal membrane between EPI/ExE and VE; RM, Reichert's membrane; the embryo is surrounded by maternal decidua, the egg cylinder is delineated with dashed lines.
- B
Immunostaining of E4.75 (top) and E5.25 (bottom) pregnant uteri cross sections, showing Fibronectin (FN1, white), Collagen IV (COLIV, white), and Laminin (LAM, white) (from left to right), GATA4 (green), and nuclei (DNA, blue). White asterisks mark the implanted embryos. Red arrowheads point at the uterine ECM.
- C
Schematic of the 3D hydrogel‐embedded embryo culture. Inset, Immunostaining of the embryos embedded and cultured 3D inside hydrogel drops until Day 2 (D2) and Day 3 (D3) showing OCT3/4 (magenta), GATA4 (green), and nuclei (DAPI, blue). White arrowheads point at the Reichert's membrane.
- D
Comparison of the epiblast (EPI) cell numbers between in utero E3.5–E5.5 embryos and embryos embedded and cultured 3D inside hydrogel drops until Days 2–3 (D2–3). n = 7 (D2) and n = 14 (D3). The midline marks the median, and the boxes indicate the interquartile range. Mann–Whitney's U test P‐value.
- E
Schematic of the embryo morphology criteria (I‐III), based on which the efficiency of the ex vivo culture is evaluated.
- E′
Immunostaining of 3E‐uterus embryos from Day 3 showing OCT3/4 (magenta), GATA4 (green), Laminin (LAM, white), and nuclei (DNA, blue). The embryos that form egg cylinder (I) show the egg cylinder axis in line with the crypt axis (II), and form Reichert's membrane (III), are considered to be successfully developed (outlined in green; 46%; n = 12 of 26, pooled from three independent experiments). White arrowheads point at Reichert's membrane.
- F
3E‐uterus efficiency for embryo culture inside cylindrical crypts with different diameters, calculated across 2 (80 μm), 3 (100 μm), 3 (120 μm), 5 (140 μm), and 3 (160 μm) independent experiments.
- G
3E‐uterus efficiency for embryo culture inside funnel‐shaped microwells made of nonbiodegradable PEG with RGD, calculated across 2 (1.5% PEG concentration), 2 (1.7%), 2 (2%), and 1 (3%) independent experiments.
- H
Immunostaining of 3E‐uterus embryos from Day 3 (D3) grown in 1.5%, 2%, 2.5%, and 7% PEG precursor concentrations (from left to right), showing OCT3/4 (magenta), GATA4 (green), Collagen IV (COLIV, white), and nuclei (DNA, blue).
- I
3E‐uterus efficiency at Day 3 at a 1.5–7% range of PEG precursor content, calculated across 3 (1.5%), 4 (1.75%), 3 (2%), 3 (2.25%), 3 (2.5%), 3 (2.75%), 3 (6%), and 2 (7%) independent experiments. Inset, rheological measurement showing linear relationship between the PEG precursor content (%, w/v) and the Shear modulus (kPa).
- J
Total cell number (EPI + VE) vs in utero developmental stage. The days of 3E‐uterus culture were matched with the in utero stages based on the log‐linear regression. Equation of the regression line for the total cell number (EPI and VE) is y = 0.133e1.489x; that for the EPI cell number is y = 0.036e1.617x. n = 6 (E3.5), n = 21 (E4.5), n = 28 (E4.75), n = 20 (E5.0), n = 20 (E5.25), n = 21 (E5.5), n = 21 (E5.75) and 22 (E6.0). Y scale, log 10.
- K
Immunostaining of E5.25 pregnant uterus cross section (left) and 3E‐uterus embryo from Day 3 (right) showing H2B‐GFP (marks the embryo in green), Cytokeratin 8 (KRT8, red), pan‐Laminin (pan‐LAM, white), and nuclei (DNA, blue). Right, 4× zoom; bottom, 2× zoom. White arrowheads point at Reichert's membrane. Scale bars, 50 μm, 100 μm (A), 12.5 μm (J, zoom‐in).

- A, B
Immunostaining of pregnant uteri cross sections at E4.75 (A) and E5.25 (B) showing mGFP (marks the embryo in green), Fibronectin 1 (FN1, white), and nuclei (DNA, blue). Red arrowheads point at the uterine ECM. (A) right, 4× zoom into the interface between trophectoderm (TE), the uterine epithelium (E), and stroma (S). (B), In utero length (l) and diameter (d) of E5.25 embryos.
- C
Schematic of peri‐implantation embryo culture. Embryos are recovered at E3.5, treated with Tyrode's solution to remove Zona pellucida (ZP), and embedded into the crypts on the day of recovery (D0). IVC1 and ICV2 stand for “In Vitro Culture” medium 1 and 2, respectively. Inset, schematic of the hydrogel composition. 8‐arm Poly(ethylene glycol) (PEG, gray) molecules, cross‐linked via metalloprotease‐cleavable peptides (Peptide, green), and functionalized with RGDSPG peptide (Arg‐Gly‐Asp‐Ser‐Pro‐Gly, “RGD,” red).
- D, E
Immunostaining of in utero E4.5 and E5.25 embryos (D) and 3E‐uterus Day 2 and Day 3 embryos (E) showing OCT3/4 (magenta), GATA4 (green), and nuclei (DAPI, blue).
- F
Numbers of epiblast (EPI) cells (x‐axis) vs visceral endoderm (VE) cells (y‐axis) that cover epiblast (shown on the bottom right scheme) in the embryos developed in utero until E5.5 (Ichikawa et al, ; E3.5–E5.5) and the embryos successfully developed by 3E‐uterus until Day 3 (D1–3). n = 5 (E3.5), n = 21 (E4.5), n = 28 (E4.75), n = 20 (E5.0), n = 20 (E5.25), n = 21 (E5.5); n = 20, two replicates pooled (D1), n = 13 of 28, three replicates pooled (D2), n = 12 of 26, three replicates pooled (D3). X/Y scale, log 10, arrows point to the representative 3E‐uterus embryos shown in (E).
- G
Left to right, Egg cylinder's length, diameter, and the length‐to‐diameter ratio between embryos developed in utero until E5.25 and 3E‐uterus embryos from Day 3 (D3). n = 14 and 12, respectively. Data points correspond to individual embryos, midline marks the median, and boxes indicate interquartile range. Student's t‐test and the Mann–Whitney's U test P‐values.
- H
Cell number‐based correspondence between in utero and 3E‐uterus embryo development.
- I, J
(I) Immunostaining of E4.75 pregnant uterus cross section (left) and 3E‐uterus embryo from Day 2 (right) showing H2B‐GFP (marks the embryo in green), TFAP2C (yellow), and nuclei (DNA, blue). (J) Immunostaining of pregnant uterus cross section (left, E5.25) and 3E‐uterus embryo (right, Day 3), showing H2B‐GFP (marking the embryo, green), TFAP2C (yellow), and nuclei (DNA, blue). Yellow arrowheads mark differentiated trophoblast cells. White asterisks indicate the epiblast of the implanted embryos. Data information: Scale bars, 50 μm, 12.5 μm (A, right).

- A
Single cells were collected and sequenced from 6 to 10 embryos for each experimental condition (E4.5, E5.25, Day 2, Day 3; red, yellow, light and dark blue, respectively) from two independent replicates (litters, N = 8 in total). After quality‐based filtering, in total 1,234 transcriptomes were used for further analysis.
- B
The UMAP of single‐cell transcriptomes colored by the in utero and 3E‐uterus experimental conditions shown on the same graph: left, E4.5 (red) and D2 (light blue); right, E5.25 (yellow) and D3 (dark blue).
- C
The UMAP colored by the clustering outcome (Leiden, Traag et al, 2019), identifying epiblast (EPI, pink; n = 421), visceral and parietal endoderm (VE/PE, green; n = 421), polar trophectoderm/extraembryonic ectoderm (pTE/ExE, gray; n = 197), and mural trophectoderm/trophoblast (mTE/TB, blue; n = 195) cells. Bottom, percentage of the identified cell types across the experimental conditions.
- D–G
UMAPs for in utero (top, n = 566) and 3E‐uterus (bottom, n = 668) cells colored by the normalized gene expression of Oct3/4 and Sox2 (EPI, D), Esrrb and Cdx2 (pTE/ExE, E), Gata4 and Dab2 (VE/PE, F), Tfap2c and Gata2 (mTE/TB, G).
- H
Dotplot showing quantification of the gene expression within the cell groups arranged by the experimental condition (E4.5, E5.25, D2, and D3) and cell type (EPI, magenta; VE/PE, green; mTE/TB, blue; pTE/ExE, gray), corresponding to y‐axis. The normalized gene expression level is denoted by the color of each dot, along with the fraction of the cell number in the group where marker gene expression was detected (dot size). X‐axis shows marker gene names. The plot indicates lineage‐specific expression of the marker genes and comparable gene expression levels between in utero (E4.5 and E5.25) and 3E‐uterus (Day 2 and Day 3) conditions across different cell types (EPI, magenta; VE/PE, green; mTE/TB, blue; pTE/ExE, gray).
- I
Heatmap of unsupervised clustering of all in utero (black) and all 3E‐uterus (white) cells based on Pearson correlation using the first 50 principal component values from the expression of all protein‐coding genes. The plot shows that the cells from in utero and 3E‐uterus cluster predominantly based on the cell type rather than the sample origin.

- A–D
UMAPs colored by the normalized expression of epiblast (A, EPI), polar trophectoderm/extraembryonic ectoderm (B, pTE/ExE), visceral, anterior visceral, and parietal endoderm (C, VE/AVE/PE), and mural trophectoderm/trophoblast (D, mTE/TB) across In utero (top, n = 566) and 3E‐uterus (bottom, n = 668) cells.
- E
The UMAP colored by the experimental conditions: 3E‐uterus (D2, light blue; D3, dark blue) and in utero (E.4.5, red; E5.25, yellow), total n = 1,234.
- F
The numbers of single‐cell transcriptomes per experimental condition and cell type.
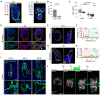
- A
Immunostaining of embryos cultured in 3E‐uterus with RGD (left) and without RGD (right) for 3 days showing pan‐Laminin (pan‐LAM, white), GATA4 (green), and nuclei (DNA, blue). Yellow arrowheads point at the Reichert's membrane.
- B
Developmental efficiency of 3E‐uterus with RGD and without RGD. Dots correspond to efficiency values in experimental replicates (N = 5 and 3, respectively). Error bars mark SD, Student's t‐test P‐value.
- C
Numbers of epiblast (OCT4+, EPI) and primitive endoderm (GATA4+, PrE) cells in all embryos grown in 3E‐uterus with RGD (n = 19, pooled from three biological replicates) and without RGD (n = 25, pooled from three biological replicates) at Day 3 of 3E‐uterus. The midline marks the median, the boxes indicate the interquartile range, and the whiskers extend maximum ± 1.5x interquartile range.
- D
Immunostaining of 3E‐uterus embryo from Day 2 showing integrin beta 1 (ITGB1, green), active ITGB1 (12G10, red), and nuclei (DNA, blue). Bottom, 2× zoom. White arrowheads point at the apical surface of the trophoblast (TB) cells.
- E–H
(E, G) Immunofluorescence of the embryo grown in utero until E4.5 and 3E‐uterus embryo from Day 2 (G) showing ZO‐1 (green), phosphor‐Ezrin/Radixin/Moesin (pERM, red), and nuclei (DNA, blue). Right, 4x zoom into the mural TE (mTE) cell. (F, H) Corresponding intensity profile plots for ZO1 and pERM signals along the cell surface outlined in (E) and (G), right.
- I
left to right, Immunostaining of the E4.75, E5.0, and E5.25 pregnant uteri cross sections showing Lifeact‐GFP (marking the embryos in green) and nuclei (DNA, blue). Bottom, 4× zoom. White arrowheads point at the mural trophectoderm (mTE)/TB membrane protrusions. White asterisks indicate the epiblast of the implanted embryos.
- J
Time‐lapse images of the developing Lifeact‐GFP (green);mTmG (magenta) embryo. The crypt surface is outlined. t = 00:00, hours: minutes from recovery at E3.5.

- A, B
Immunofluorescence of E4.5 embryos showing nuclei (DNA, blue), integrin beta 1 (ITGB1, red) (A), and active ITGB1 (12G10, green) (B). Right, 4× zoom‐ins. Arrowheads point to the apicobasal integrin localization in mural TE.
- C
Immunofluorescence of the blastocyst‐stage embryo (E3.5) showing integrin beta 1 (ITGB1, red), and nuclei (DAPI, blue). Right, 4× zoom. Arrowheads point to the basal integrin localization in TE.
- D
3D projections of time‐lapse images of the developing ZO1‐GFP (green);mTmG (magenta) embryo. Bottom, 2.5× zoom into the TE cell; white arrowheads mark cell–cell interfaces.
- E
Immunofluorescence of the 3E‐uterus embryo after live imaging, simultaneously stained for ZO1‐GFP (green) PARD6B (red), and nuclei (DNA, blue). From left to right, ZO1‐GFP, PARD6B, composite image channels. Bottom, 4× zoom of the TB cell.
- F
Intensity profile of ZO1 and PARD6B signals along the cell surface outlined in (E, bottom), including apical and basolateral regions.
- G
Time‐lapse images of the developing Myh9‐GFP (green);mTmG (magenta) embryo. The crypt surface is outlined.
- H
Immunofluorescence of the 3E‐uterus embryo after live imaging showing Myh9‐GFP (green) phosphor‐MLC (T18/S19) (red), and nuclei (DNA, blue). Bottom, 2× zoom. White arrowheads point at the apical TB cell surface.
- I
Immunofluorescence of Day 3 3E‐uterus embryo, showing maximum Z‐projection of F‐actin signal (white). Bottom, 2× zoom; right, 4× zoom. Yellow arrows mark trophoblast cell membrane protrusions. Invasive trophoblast cell protrusions at least 10 μm deep inside the biodegradable LDTM PEG matrix are consistently observed in 86% of all WT embryos at the Day 3 of 3E‐uterus.
- J, K
immunofluorescence of the mural TE (mTE) cell of the embryo grown in utero until E4.5 (left) and 3E‐uterus embryo from Day 2 (right) showing ZO‐1 (green), phosphor‐Ezrin/Radixin/Moesin (pERM, red), and nuclei (DNA, blue) without the outline, corresponding to Fig 3E and G. t = 00:00, Hours: Minutes from recovery at E3.5. Scale bars, 50 μm, 25 μm (2× zoom), 20 μm (2.5× zoom), 12.5 μm (4× zoom).
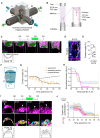
Schematic of the MuVi‐SPIM setup with two low‐NA illumination objective lenses (IL), two high‐NA imaging objective lenses (IM), and the controlled environmental imaging chamber with the sample holder (yellow Arrow).
Schematic of the sample holder. The outer FEP tube (∅1.8 mm, l = 25 mm) is mounted on top of the sealed glass capillary and filled with IVC medium. The inner FEP tube (∅1 mm, l = 3 mm) contains the crypt and is supported by the PDMS holder from the bottom. The embryo is mounted from the top. The outer FEP tube is closed with the PDMS cap with ∅0.6 mm opening for the gas exchange.
Time‐lapse images of the mural TE (mTE) in the mTmG (magenta) developing embryo. The fitted droplet model (embryo) and the frustum shape (crypt) are in green; an exemplar contact angle (θ) between mTE and the crypt surface is shown. Apical and basal sides of mural TE are marked with arrows.
Immunostaining of the E4.75 pregnant uterus cross section showing mGFP (marks the embryo in magenta), Fibronectin 1 (FN1, white), and nuclei (DNA, blue). An exemplar contact angle between mTE and the uterine basal membrane is shown.
In utero contact angle values from six E4.75 embryos collected from three biological replicates, measured in 1–2 cross sections. The midline marks the median, the boxes indicate the interquartile range, and the whiskers extend maximum ± 1.5× interquartile range.
Schematic of the active droplet in a frustum‐shaped confinement; θa and θb denote top and bottom contact angles, respectively.
Simulated contact angle dynamics for constant tension (dashed line) and decreasing tension (solid line) with experimental data (in orange). Error bars denote SEM. See also Fig EV4H and Appendix Fig S4.
Inferred dynamics of the normalized embryo‐substrate interfacial tension difference. Colors correspond to independent experiments.
Top, time‐lapse images of the polar TE (pTE) in mTmG (magenta) developing embryo. Exemplar pTE cells are marked with arrowheads, cell perimeter is outlined. Middle, corresponding images of the 3D cell membrane segmentation. Bottom, the schematic of pTE cell columnarization and invagination. Apical side of polar TE is marked with an arrow.
Dynamics of the width‐to‐height aspect ratio of the pTE cells. Colors correspond to independent experiments (same as H). Average values across 15–20 cells per time point (solid line) and standard deviations (shaded area) are shown. t = 00:00, hours: minutes from recovery at E3.5.

The equilibrium shape of the droplet in a cylindrical confinement of radius r is described by the distance h between the two contact lines and by the height y of the top and bottom spherical caps, corresponding to polar and mural TE, respectively. These caps can also be characterized by the curvature radius R and angle φ. The contact angle θ depends on the droplet–medium tension γ 0 and the Young tension Δγ.
The droplet in a conical frustum with angle α is described by the positions of the top and bottom contact lines z 1, z 2 measured from the conical tip z 0 = 0, and by the heights of the top and bottom spherical caps z 3 and z 4, respectively. When the caps curve into the embryo, their heights assume negative values.
Bifurcation diagram for the equilibrium solutions Equation S6 (Appendix) of the droplet in cylindrical confinement. The solid line corresponds to the stable solution y −, whereas the dashed line denotes the unstable branch y +.
Top, Calculated equilibrium shapes of the droplet in cylindrical confinement at the transition to total wetting (I), in the regime of partial wetting (II, III), and dewetting (IV). Bottom, Time‐lapse images of mTmG signal (magenta) in the embryos growing in 3E‐uterus with RGD, corresponding to the I‐III wetting regimes and without RGD, corresponding to dewetting (IV). T = 00:00, hours: minutes after recovery at E3.5. The crypt surface is outlined.
Sigmoid model of the Young tension adaptation Equation S16 drawn for three values of the modulation parameter a ≥ 0. Constants c 1 and c 2 specify the initial and final values of the normalized tension. The adaptation begins at a time instance t 1 and ends at a time instance t 2. A full specification of the model requires five constants, for example, the mid‐time t 0 = (t 1 + t 2)/2, the duration Δt = t 2 − t 1, the constants c 1 and c 2, and the modulation parameter a.
Volume dynamics in the developing embryos between 36 and 56 h after E3.5. Colors correspond to different embryos; n = 3.
Contact angle (θb) dynamics in developing embryos. Colors correspond to different embryos imaged in time intervals between 20 and 72 h after E3.5; n = 10.
From left to right, simulated contact angle dynamics for constant tension (dashed line), and decreasing tension (solid line), with experimental data points (green and red points for θa and θb, respectively) for three different embryos between 36 and 56 h from recovery at E3.5. Error bars denote SEM. Scale bar, 50 μm.

Time‐lapse images of the developing CDX2‐GFP (green); mTmG (magenta) embryo. t = 00:00, hours: minutes after recovery at E3.5.
Quantification of the ExE cell numbers for three independent experiments.
Time‐lapse images of the H2B‐GFP (green); mTmG (magenta) developing embryo. Bottom, 2x zoom into the epiblast region. Right and bottom, YZ and XZ image sections show 3D resolution. Yellow arrows indicate egg cylinder growth; asterisk, the pro‐amniotic cavity formation.
Egg cylinder length between 50 and 68 h after recovery at E3.5. Colors correspond to independent experiments.
Epiblast cell lineage dendrograms. Right, corresponding cells marked as dots with different colors overlaying the dendrograms and the image slices; cell lineage tracks are depicted as a 2D overlay.
An increase in epiblast tissue volume between 56 and 64 h after recovery at E3.5. Colors correspond to independent experiments (same as D).

- A
Schematic of the hypothesis that adhesion‐induced migration of trophoblast (TB) cells generates space for embryo growth.
- B
3D projections of time‐lapse images of the developing H2B‐GFP (green) embryo. Trajectories of individual mural TE (mTE) cells are marked with red lines. The crypt surface is outlined.
- C
Trajectories of mTE cells in an XY plane, normalized to the starting coordinates. End coordinates are marked with red dots; n = 29.
- D
Displacement of mTE cells along the Y‐axis in relation to imaging time; n = 29. The linear regression fit is shown in black, y = −9.76‐2.71x, R 2 = 0.41.
- E
Time‐lapse images of the developing mTmG (gray) embryo in the Z plane corresponding to the crypt surface. The arrow indicates direction of migration.
- F
Left to right, Immunostaining of WT, Rac1 +/−, and Rac1 −/− embryos, cultured up to Day 3 (D3) in 3E‐uterus, showing OCT3/4 (magenta), pan‐Laminin (pan‐LAM, white), F‐actin (yellow), and nuclei (DNA, blue). The crypt surface is outlined, and the arrow points at the invasive trophoblast cell protrusion.
- G, H
Embryo length and epiblast cell number in WT, Rac1 +/−, and Rac1 −/− embryos. n = 17 (WT), 23 (Rac1 +/−), 13 (Rac1 −/−), embryos pooled from five experimental replicates (epiblast cell number) and n = 22 (WT), 30 (Rac1 +/−), 18 (Rac1 −/−), embryos pooled from seven experimental replicates (embryo length). Data points correspond to embryos; the midline marks the median, the boxes indicate the interquartile range, and the whiskers extend maximum ± 1.5× interquartile range; the red dots mark representative embryos shown in (F). Mann–Whitney's U test P‐value.
- I, J
Egg cylinder elongation and RM movement in a downward (I) and upward (J) embryo orientations. (I, J) left, Schematic of the egg cylinder tip (magenta) and the Reichert's Membrane (RM, blue) movement within an experimentally controlled space (green). Coordinates are scaled to the starting coordinate of the egg cylinder's tip. (I, J) middle, Time‐lapse images of H2B‐GFP (green); mTmG (magenta) developing embryos. (I, J) right, Movement of the egg cylinder tip (magenta) and RM (blue) along the crypt axis. Solid lines and shaded regions indicate average and SD values across two (downward) and four (upward) independent experiments. t = 00:00, hours: minutes from recovery at E3.5.

Directionality of the mTE/TB migration along the X, Y, and Z axes (green, red, and blue, respectively) between subsequent hours of live imaging. n = 29.
Left, Mural TE (mTE) cell trajectories for three different embryos; coordinates in XY plane are normalized to the starting coordinates. End coordinates are marked with red dots. Right, Displacement of mTE cells along the Y‐axis vs imaging time post‐E3.5. From top to the bottom, n = 61, 58, 51, respectively. The linear regression fit is shown as a black line.
Directionality of the mTE/TB migration along the X, Y, and Z axes (green, red, and blue, respectively) between subsequent hours of live imaging for three embryos (from top to bottom). n = 61, 58, 51, respectively.
Distribution density of the average TB velocities (μm/h). n = 255, pooled from six embryos.
TB migration speed (μm/h) vs imaging time post‐E3.5. Colors correspond to the three embryos from (B) and (C).
Persistence of the nearest mTE/TB four‐cell neighborhood between subsequent hours of live imaging.

References
-
- Alert R, Casademunt J (2019) Role of substrate stiffness in tissue spreading: wetting transition and tissue durotaxis. Langmuir 35: 7571–7577 - PubMed
-
- Andreotti B, Snoeijer JH (2020) Statics and dynamics of soft wetting. Annu Rev Fluid Mech 52: 285–308
-
- Bedzhov I, Leung CY, Bialecka M, Zernicka‐Goetz M (2014) In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 9: 2732–2739 - PubMed
-
- Behringer R, Gertsenstein M, Nagy K, Nagy A (2014) Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press;

