Noncanonical HPV carcinogenesis drives radiosensitization of head and neck tumors
- PMID: 37523561
- PMCID: PMC10410762
- DOI: 10.1073/pnas.2216532120
Noncanonical HPV carcinogenesis drives radiosensitization of head and neck tumors
Abstract
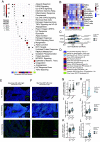
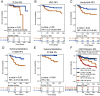
We analyzed transcriptional data from 104 HPV+ (Human papillomavirus) HNSCC (head and neck squamous cell carcinoma) tumors together with two publicly available sources to identify highly robust transcriptional programs (modules) which could be detected consistently despite heterogeneous sequencing and quantification methodologies. Among 22 modules identified, we found a single module that naturally subclassifies HPV+ HNSCC tumors based on a bimodal pattern of gene expression, clusters all atypical features of HPV+ HNSCC biology into a single subclass, and predicts patient outcome in four independent cohorts. The subclass-defining gene set was strongly correlated with Nuclear factor kappa B (NF-κB) target expression. Tumors with high expression of this NF-κB module were rarely associated with activating PIK3CA alterations or viral integration, and also expressed higher levels of HPHPV E2 and had decreased APOBEC mutagenesis. Alternatively, they harbored inactivating alterations of key regulators of NF-κB, TNF receptor associated factor 3 (TRAF3), and cylindromatosis (CYLD), as well as retinoblastoma protein (RB1). HPV+ HNSCC cells in culture with experimental depletion of TRAF3 or CYLD displayed increased expression of the subclass-defining genes, as well as robust radio-sensitization, thus recapitulating both the tumor transcriptional state and improved treatment response observed in patient data. Across all gene sets investigated, methylation to expression correlations were the strongest for the subclass-defining, NF-κB-related genes. Increased tumor-infiltrating CD4+ T cells and increased Estrogen receptors alpha (ERα) expression were identified in NF-κB active tumors. Based on the relatively high rates of cure in HPV+ HNSCC, deintensification of therapy to reduce treatment-related morbidity is being studied at many institutions. Tumor subclassification based on oncogenic subtypes may help guide the selection of therapeutic intensity or modality for patients with HPV+ HNSCC.
Keywords: HPV; head and neck cancer; patient outcome; radiosensitization; tumor microenvironment.
Conflict of interest statement
T.P.S., W.G.Y., and N.I., hold intellectual property related to the presented work. UNC 21-0070 reporting filing of PCT application No. PCT/US22/32871; OLG Ref. 150-34-PCT.
Figures






References
-
- Viens L. J., et al. , Human papillomavirus-associated cancers–United States, 2008–2012. MMWR Morb. Mortal Wkly. Rep. 65, 661–666 (2016). - PubMed
-
- Shiboski C. H., Schmidt B. L., Jordan R. C. K., Tongue and tonsil carcinoma: Increasing trends in the U.S. population ages 20–44 years. Cancer 103, 1843–1849 (2005). - PubMed
-
- Burtness B., et al. , Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 394, 1915–1928 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 DE030707/DE/NIDCR NIH HHS/United States
- U24 CA196172/CA/NCI NIH HHS/United States
- U10CA180820/CA/NCI NIH HHS/United States
- K08 DE029241/DE/NIDCR NIH HHS/United States
- R01 DE027942/DE/NIDCR NIH HHS/United States
- UG1CA233337/CA/NCI NIH HHS/United States
- U10 CA180820/CA/NCI NIH HHS/United States
- U10 CA180794/CA/NCI NIH HHS/United States
- R01 DE031297/DE/NIDCR NIH HHS/United States
- U10CA180794/CA/NCI NIH HHS/United States
- KO8-DE029241/DE/NIDCR NIH HHS/United States
- R01DE031297/DE/NIDCR NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- U01 DE029754/DE/NIDCR NIH HHS/United States
- R01 DE026471/DE/NIDCR NIH HHS/United States
- R01DE027942/DE/NIDCR NIH HHS/United States
- DE030707/DE/NIDCR NIH HHS/United States
- U01DE029754/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

