A microfluidic biosensor architecture for the rapid detection of COVID-19
- PMID: 37524456
- PMCID: PMC10251744
- DOI: 10.1016/j.aca.2023.341378
A microfluidic biosensor architecture for the rapid detection of COVID-19
Abstract
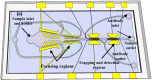
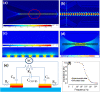
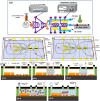
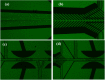
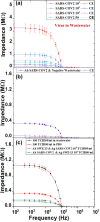
The lack of enough diagnostic capacity to detect severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) has been one of the major challenges in the control the 2019 COVID pandemic; this led to significant delay in prompt treatment of COVID-19 patients or accurately estimate disease situation. Current methods for the diagnosis of SARS-COV-2 infection on clinical specimens (e.g. nasal swabs) include polymerase chain reaction (PCR) based methods, such as real-time reverse transcription (rRT) PCR, real-time reverse transcription loop-mediated isothermal amplification (rRT-LAMP), and immunoassay based methods, such as rapid antigen test (RAT). These conventional PCR methods excel in sensitivity and specificity but require a laboratory setting and typically take up to 6 h to obtain the results whereas RAT has a low sensitivity (typically at least 3000 TCID50/ml) although with the results with 15 min. We have developed a robust micro-electro-mechanical system (MEMS) based impedance biosensor fit for rapid and accurate detection of SARS-COV-2 of clinical samples in the field with minimal training. The biosensor consisted of three regions that enabled concentrating, trapping, and sensing the virus present in low quantities with high selectivity and sensitivity in 40 min using an electrode coated with a specific SARS-COV-2 antibody cross-linker mixture. Changes in the impedance value due to the binding of the SARS-COV-2 antigen to the antibody will indicate positive or negative result. The testing results showed that the biosensor's limit of detection (LoD) for detection of inactivated SARS-COV-2 antigen in phosphate buffer saline (PBS) was as low as 50 TCID50/ml. The biosensor specificity was confirmed using the influenza virus while the selectivity was confirmed using influenza polyclonal sera. Overall, the results showed that the biosensor is able to detect SARS-COV-2 in clinical samples (swabs) in 40 min with a sensitivity of 26 TCID50/ml.
Keywords: Biosensor; Dielectrophoresis; Impedance measurement; Microfluidic; Polymerase chain reaction (PCR); SARS-COV-2; Trapping and focusing electrodes.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Wang H., Paulson K.R., Pease S.A., Watson S., Comfort H., Zheng P., Aravkin A.Y., Bisignano C., Barber R.M., Alam T., Fuller J.E., et al. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020–21. Lancet. 2022;399(10334):1513–1536. doi: 10.1016/S0140-6736(21)02796-3. - DOI - PMC - PubMed
-
- World Health Organization "WHO coronavirus (COVID19) dashboard coronavirus (COVID-19) https://covid19.who.int/ Dashboard [Internet].[consultado 25 Sep 2021].".
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

