An impaired splicing program underlies differentiation defects in hSOD1G93A neural progenitor cells
- PMID: 37524863
- PMCID: PMC11072603
- DOI: 10.1007/s00018-023-04893-7
An impaired splicing program underlies differentiation defects in hSOD1G93A neural progenitor cells
Abstract
Amyotrophic lateral sclerosis (ALS) is an adult devastating neurodegenerative disease characterized by the loss of upper and lower motor neurons (MNs), resulting in progressive paralysis and death. Genetic animal models of ALS have highlighted dysregulation of synaptic structure and function as a pathogenic feature of ALS-onset and progression. Alternative pre-mRNA splicing (AS), which allows expansion of the coding power of genomes by generating multiple transcript isoforms from each gene, is widely associated with synapse formation and functional specification. Deciphering the link between aberrant splicing regulation and pathogenic features of ALS could pave the ground for novel therapeutic opportunities. Herein, we found that neural progenitor cells (NPCs) derived from the hSOD1G93A mouse model of ALS displayed increased proliferation and propensity to differentiate into neurons. In parallel, hSOD1G93A NPCs showed impaired splicing patterns in synaptic genes, which could contribute to the observed phenotype. Remarkably, master splicing regulators of the switch from stemness to neural differentiation are de-regulated in hSOD1G93A NPCs, thus impacting the differentiation program. Our data indicate that hSOD1G93A mutation impacts on neurogenesis by increasing the NPC pool in the developing mouse cortex and affecting their intrinsic properties, through the establishment of a specific splicing program.
Keywords: ALS; Alternative splicing; Neural progenitor cells.
© 2023. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures
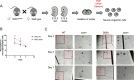

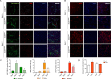
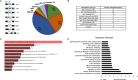

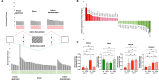
References
-
- Avossa D, Grandolfo M, Mazzarol F, Zatta M, Ballerini L. Early signs of motoneuron vulnerability in a disease model system: characterization of transverse slice cultures of spinal cord isolated from embryonic ALS mice. Neuroscience. 2006;138:1179–1194. doi: 10.1016/j.neuroscience.2005.12.009. - DOI - PubMed
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

