Isothermal self-assembly of multicomponent and evolutive DNA nanostructures
- PMID: 37524905
- PMCID: PMC10656289
- DOI: 10.1038/s41565-023-01468-2
Isothermal self-assembly of multicomponent and evolutive DNA nanostructures
Abstract
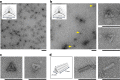
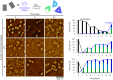
Thermal annealing is usually needed to direct the assembly of multiple complementary DNA strands into desired entities. We show that, with a magnesium-free buffer containing NaCl, complex cocktails of DNA strands and proteins can self-assemble isothermally, at room or physiological temperature, into user-defined nanostructures, such as DNA origamis, single-stranded tile assemblies and nanogrids. In situ, time-resolved observation reveals that this self-assembly is thermodynamically controlled, proceeds through multiple folding pathways and leads to highly reconfigurable nanostructures. It allows a given system to self-select its most stable shape in a large pool of competitive DNA strands. Strikingly, upon the appearance of a new energy minimum, DNA origamis isothermally shift from one initially stable shape to a radically different one, by massive exchange of their constitutive staple strands. This method expands the repertoire of shapes and functions attainable by isothermal self-assembly and creates a basis for adaptive nanomachines and nanostructure discovery by evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Shaken, not heated: DNA self-assembly at room temperature.Nat Nanotechnol. 2023 Nov;18(11):1257-1258. doi: 10.1038/s41565-023-01467-3. Nat Nanotechnol. 2023. PMID: 37550575 No abstract available.
References
-
- Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. - PubMed
-
- Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. - PubMed
-
- Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. - PubMed
-
- Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 258782/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- ANR-18-CE06-0019/Agence Nationale de la Recherche (French National Research Agency)
- ANR-18-CE07-0001/Agence Nationale de la Recherche (French National Research Agency)
- Summer Program/MEXT | Japan Society for the Promotion of Science (JSPS)
- Overseas Research Fellowship/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources

