Garcinolic Acid Distinguishes Between GACKIX Domains and Modulates Interaction Networks
- PMID: 37525583
- PMCID: PMC10870240
- DOI: 10.1002/cbic.202300439
Garcinolic Acid Distinguishes Between GACKIX Domains and Modulates Interaction Networks
Abstract
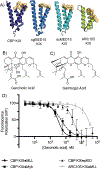
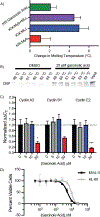
Natural products are often uniquely suited to modulate protein-protein interactions (PPIs) due to their architectural and functional group complexity relative to synthetic molecules. Here we demonstrate that the natural product garcinolic acid allosterically blocks the CBP/p300 KIX PPI network and displays excellent selectivity over related GACKIX motifs. It does so via a strong interaction (KD 1 μM) with a non-canonical binding site containing a structurally dynamic loop in CBP/p300 KIX. Garcinolic acid engages full-length CBP in the context of the proteome and in doing so effectively inhibits KIX-dependent transcription in a leukemia model. As the most potent small-molecule KIX inhibitor yet reported, garcinolic acid represents an important step forward in the therapeutic targeting of CBP/p300.
Keywords: CBP/p300; acute myelogenous leukemia; cMyb; natural product; protein-protein interaction inhibitor.
© 2023 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

