Unlocking the secrets of ABEs: the molecular mechanism behind their specificity
- PMID: 37526140
- PMCID: PMC10586758
- DOI: 10.1042/BST20221508
Unlocking the secrets of ABEs: the molecular mechanism behind their specificity
Abstract
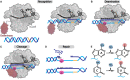
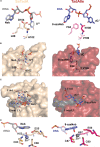
CRISPR-Cas, the bacterial immune systems, have transformed the field of genome editing by providing efficient, easily programmable, and accessible tools for targeted genome editing. DNA base editors (BE) are state-of-the-art CRISPR-based technology, allowing for targeted modifications of individual nucleobases within the genome. Among the BEs, adenine base editors (ABEs) have shown great potential due to their ability to convert A-to-G with high efficiency. However, current ABEs have limitations in terms of their specificity and targeting range. In this review, we provide an overview of the molecular mechanism of ABEs, with a focus on the mechanism of deoxyadenosine deamination by evolved tRNA-specific adenosine deaminase (TadA). We discuss how mutations and adjustments introduced via both directed evolution as well as rational design have improved ABE efficiency and specificity. This review offers insights into the molecular mechanism of ABEs, providing a roadmap for future developments in the precision genome editing field.
Keywords: DNA adenine base editor; DNA base editing; DNA deaminase; deamination; genome editing; tRNA deaminase.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

References
-
- Stafuzza, N.B., Zerlotini, A., Lobo, F.P., Yamagishi, M.E.B., Chud, T.C.S., Caetano, A.R.et al. (2017) Single nucleotide variants and InDels identified from whole-genome re-sequencing of Guzerat, Gyr, Girolando and Holstein cattle breeds. PLoS ONE 12, e0173954 10.1371/journal.pone.0173954 - DOI - PMC - PubMed

