Peripheral Blood DNA Methylation Signatures and Response to Tofacitinib in Moderate-to-severe Ulcerative Colitis
- PMID: 37526299
- PMCID: PMC11324342
- DOI: 10.1093/ecco-jcc/jjad129
Peripheral Blood DNA Methylation Signatures and Response to Tofacitinib in Moderate-to-severe Ulcerative Colitis
Abstract
Introduction: Predictive biomarkers for treatment efficacy of ulcerative colitis [UC] treatments are lacking. Here, we performed a longitudinal study investigating the association and potential predictive power of genome-wide peripheral blood [PB] DNA methylation signatures and response to tofacitinib treatment in UC.
Methods: We recruited moderate-to-severe UC patients starting tofacitinib treatment, and measured PB DNA methylation profiles at baseline [T1], after 8 weeks [T2], and in a subset [n = 8] after a median of 20 weeks [T3] using the Illumina Infinium HumanMethylation EPIC BeadChip. After 8 weeks, we distinguished responders [R] from non-responders [NR] based on a centrally read endoscopic response [decrease in endoscopic Mayo score ≥1 or Ulcerative Colitis Endoscopic Index of Severity ≥2] combined with corticosteroid-free clinical and/or biochemical response. T1 PB samples were used for biomarker identification, and T2 and publicly available intraclass correlation [ICC] data were used for stability analyses. RNA-sequencing was performed to understand the downstream effects of the predictor CpG loci.
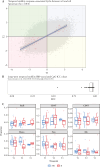
Results: In total, 16 R and 15 NR patients, with a median disease duration of 7 [4-12] years and overall comparable patient characteristics at baseline, were analysed. We identified a panel of 53 differentially methylated positions [DMPs] associated with response to tofacitinib [AUROC 0.74]. Most DMPs [77%] demonstrated both short- and long-term hyperstability [ICC ≥0.90], irrespective of inflammatory status. Gene expression analysis showed lower FGFR2 [pBH = 0.011] and LRPAP1 [pBH = 0.020], and higher OR2L13 [pBH = 0.016] expression at T1 in R compared with NR.
Conclusion: Our observations demonstrate the utility of genome-wide PB DNA methylation signatures to predict response to tofacitinib.
Keywords: Epigenetics; biomarkers; personalised medicine.
© The Author(s) 2023. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
VJ, AYFLY, SvG, IH, EL, PL, PH, ML, and GD: none to declare. WdJ is involved in a company AIBiomicsBV and received speaker fees from Alimentiv, Janssen, and HoraizonBV over the past 2 years.
Figures



Comment in
-
Epigenetic Fingerprints in IBD: From Methylation Patterns to Clinical Implications.J Crohns Colitis. 2024 Aug 14;18(8):1175-1176. doi: 10.1093/ecco-jcc/jjae086. J Crohns Colitis. 2024. PMID: 38885255 No abstract available.
References
-
- Danese S, Fiocchi C.. Ulcerative colitis. N Engl J Med 2011;365:1713–25. - PubMed
-
- Roda G, Dal Buono A, Argollo M, Danese S.. JAK selectivity: more precision less troubles. Expert Rev Gastroenterol Hepatol 2020;14:789–96. - PubMed
-
- Salas A, Hernandez-Rocha C, Duijvestein M, et al.. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:323–37. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

