Targeting Alpha-Ketoglutarate Disruption Overcomes Immunoevasion and Improves PD-1 Blockade Immunotherapy in Renal Cell Carcinoma
- PMID: 37526345
- PMCID: PMC10520657
- DOI: 10.1002/advs.202301975
Targeting Alpha-Ketoglutarate Disruption Overcomes Immunoevasion and Improves PD-1 Blockade Immunotherapy in Renal Cell Carcinoma
Abstract
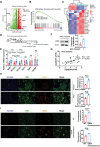
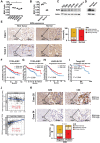
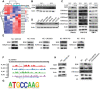
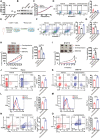
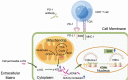
The Warburg effect-related metabolic dysfunction of the tricarboxylic acid (TCA) cycle has emerged as a hallmark of various solid tumors, particularly renal cell carcinoma (RCC). RCC is characterized by high immune infiltration and thus recommended for immunotherapeutic interventions at an advanced stage in clinical guidelines. Nevertheless, limited benefits of immunotherapy have prompted investigations into underlying mechanisms, leading to the proposal of metabolic dysregulation-induced immunoevasion as a crucial contributor. In this study, a significant decrease is found in the abundance of alpha-ketoglutarate (αKG), a crucial intermediate metabolite in the TCA cycle, which is correlated with higher grades and a worse prognosis in clinical RCC samples. Elevated levels of αKG promote major histocompatibility complex-I (MHC-I) antigen processing and presentation, as well as the expression of β2-microglobulin (B2M). While αKG modulates broad-spectrum demethylation activities of histone, the transcriptional upregulation of B2M is dependent on the demethylation of H3K4me1 in its promoter region. Furthermore, the combination of αKG supplementation and PD-1 blockade leads to improved therapeutic efficacy and prolongs survival in murine models when compared to monotherapy. Overall, the findings elucidate the mechanisms of immune evasion in anti-tumor immunotherapies and suggest a potential combinatorial treatment strategy in RCC.
Keywords: B2M; PD-1 blockade; alpha-ketoglutarate; immunoevasion; renal cell carcinoma.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Notarangelo G., Spinelli J. B., Perez E. M., Baker G. J., Kurmi K., Elia I., Stopka S. A., Baquer G., Lin J. R., Golby A. J., Joshi S., Baron H. F., Drijvers J. M., Georgiev P., Ringel A. E., Zaganjor E., McBrayer S. K., Sorger P. K., Sharpe A. H., Wucherpfennig K. W., Santagata S., Agar N. Y. R., Suva M. L., Haigis M. C., Science 2022, 377, 1519. - PMC - PubMed
-
- Wang X., Liu R., Qu X., Yu H., Chu H., Zhang Y., Zhu W., Wu X., Gao H., Tao B., Li W., Liang J., Li G., Yang W., Mol Cell 2019, 76, 148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
