Flindissone, a Limonoid Isolated from Trichilia prieuriana, Is an LXR Agonist
- PMID: 37526502
- PMCID: PMC10463221
- DOI: 10.1021/acs.jnatprod.3c00059
Flindissone, a Limonoid Isolated from Trichilia prieuriana, Is an LXR Agonist
Abstract
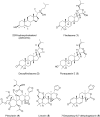
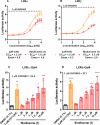
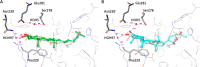
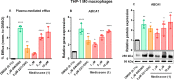
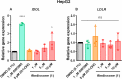
In this study, the ability of six limonoids from Trichilia prieuriana (Meliaceae) to activate the liver X receptor (LXR) was assessed. One of these limonoids, flindissone, was shown to activate LXR by reporter-gene assays. Flindissone is a ring-intact limonoid, structurally similar to sterol-like LXR ligands. In endogenous cellular settings, flindissone showed an activity profile that is characteristic of LXR agonists. It induced cholesterol efflux in THP-1 macrophages by increasing the cholesterol transporter ABCA1 and ABCG1 gene expression. In HepG2 cells, flindissone induced the expression of IDOL, an LXR-target gene that is associated with the downregulation of the LDL receptor. However, unlike synthetic and similarly to sterol-based LXR agonists, flindissone did not induce the expression of the SREBP1c gene, a major transcription factor regulating de novo lipogenesis. Additionally, flindissone also appeared to be able to inhibit post-translational activation of SREBP1c. The results presented here reveal a natural product as a new LXR agonist and point to an additional property of T. prieuriana and other plant extracts containing flindissone.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Lemmens R. H. M.Trichilia Prieureana A. Juss. In PROTA (Plant Resources of Tropical Africa/Ressources végétales de l’Afrique tropicale); Louppe D.; Oteng-Amoako A. A.; Brink M., Eds.; Wageningen, Netherlands, 2008.
-
- Passos M. S.; Nogueira T. S. R.; Azevedo O. de A.; Vieira M. G. C.; Terra W. da S.; Braz-Filho R.; Vieira I. J. C Phytochem. Rev. 2021, 20, 1055–1086. 10.1007/s11101-020-09737-x. - DOI
-
- Pagna J. I. M.; Mbekou I. M. K.; Tsamo A. T.; Mkounga P.; Frese M.; Stammler H.-G.; Fekam F. B.; Lenta B. N.; Sewald N.; Nkengfack A. E. Zeitschrift für Naturforsch. B 2021, 76, 439–446. 10.1515/znb-2021-0057. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information

