Testing for the fitness benefits of natural transformation during community-embedded evolution
- PMID: 37526972
- PMCID: PMC10482379
- DOI: 10.1099/mic.0.001375
Testing for the fitness benefits of natural transformation during community-embedded evolution
Abstract
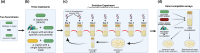
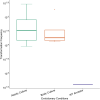
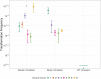
Natural transformation is a process where bacteria actively take up DNA from the environment and recombine it into their genome or reconvert it into extra-chromosomal genetic elements. The evolutionary benefits of transformation are still under debate. One main explanation is that foreign allele and gene uptake facilitates natural selection by increasing genetic variation, analogous to meiotic sex. However, previous experimental evolution studies comparing fitness gains of evolved transforming- and isogenic non-transforming strains have yielded mixed support for the 'sex hypothesis.' Previous studies testing the sex hypothesis for natural transformation have largely ignored species interactions, which theory predicts provide conditions favourable to sex. To test for the adaptive benefits of bacterial transformation, the naturally transformable wild-type Acinetobacter baylyi and a transformation-deficient ∆comA mutant were evolved for 5 weeks. To provide strong and potentially fluctuating selection, A. baylyi was embedded in a community of five other bacterial species. DNA from a pool of different Acinetobacter strains was provided as a substrate for transformation. No effect of transformation ability on the fitness of evolved populations was found, with fitness increasing non-significantly in most treatments. Populations showed fitness improvement in their respective environments, with no apparent costs of adaptation to competing species. Despite the absence of fitness effects of transformation, wild-type populations evolved variable transformation frequencies that were slightly greater than their ancestor which potentially could be caused by genetic drift.
Keywords: Acinetobacter baylyi; community evolution; evolution of sex; horizontal gene transfer; natural transformation; species interactions.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Costs and benefits of natural transformation in Acinetobacter baylyi.BMC Microbiol. 2017 Feb 15;17(1):34. doi: 10.1186/s12866-017-0953-2. BMC Microbiol. 2017. PMID: 28202049 Free PMC article.
-
Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi.ISME J. 2015 Oct;9(10):2221-31. doi: 10.1038/ismej.2015.35. Epub 2015 Apr 7. ISME J. 2015. PMID: 25848876 Free PMC article.
-
No effect of natural transformation on the evolution of resistance to bacteriophages in the Acinetobacter baylyi model system.Sci Rep. 2016 Nov 21;6:37144. doi: 10.1038/srep37144. Sci Rep. 2016. PMID: 27869203 Free PMC article.
-
Silently transformable: the many ways bacteria conceal their built-in capacity of genetic exchange.Curr Genet. 2017 Jun;63(3):451-455. doi: 10.1007/s00294-016-0663-6. Epub 2016 Nov 8. Curr Genet. 2017. PMID: 27826682 Review.
-
Steady at the wheel: conservative sex and the benefits of bacterial transformation.Philos Trans R Soc Lond B Biol Sci. 2016 Oct 19;371(1706):20150528. doi: 10.1098/rstb.2015.0528. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27619692 Free PMC article. Review.
Cited by
-
Multitasking functions of bacterial extracellular DNA in biofilms.J Bacteriol. 2024 Apr 18;206(4):e0000624. doi: 10.1128/jb.00006-24. Epub 2024 Mar 6. J Bacteriol. 2024. PMID: 38445859 Free PMC article. Review.
-
Effect of chemotherapeutic agents on natural transformation frequency in Acinetobacter baylyi.Access Microbiol. 2024 Jul 10;6(7):000733.v4. doi: 10.1099/acmi.0.000733.v4. eCollection 2024. Access Microbiol. 2024. PMID: 39135654 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

