LATE-NC risk alleles (in TMEM106B, GRN, and ABCC9 genes) among persons with African ancestry
- PMID: 37528055
- PMCID: PMC10440720
- DOI: 10.1093/jnen/nlad059
LATE-NC risk alleles (in TMEM106B, GRN, and ABCC9 genes) among persons with African ancestry
Abstract
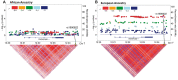
Limbic-predominant age-related TDP-43 encephalopathy (LATE) affects approximately one-third of older individuals and is associated with cognitive impairment. However, there is a highly incomplete understanding of the genetic determinants of LATE neuropathologic changes (LATE-NC) in diverse populations. The defining neuropathologic feature of LATE-NC is TDP-43 proteinopathy, often with comorbid hippocampal sclerosis (HS). In terms of genetic risk factors, LATE-NC and/or HS are associated with single nucleotide variants (SNVs) in 3 genes-TMEM106B (rs1990622), GRN (rs5848), and ABCC9 (rs1914361 and rs701478). We evaluated these 3 genes in convenience samples of individuals of African ancestry. The allele frequencies of the LATE-associated alleles were significantly different between persons of primarily African (versus European) ancestry: In persons of African ancestry, the risk-associated alleles for TMEM106B and ABCC9 were less frequent, whereas the risk allele in GRN was more frequent. We performed an exploratory analysis of data from African-American subjects processed by the Alzheimer's Disease Genomics Consortium, with a subset of African-American participants (n = 166) having corroborating neuropathologic data through the National Alzheimer's Coordinating Center (NACC). In this limited-size sample, the ABCC9/rs1914361 SNV was associated with HS pathology. More work is required concerning the genetic factors influencing non-Alzheimer disease pathology such as LATE-NC in diverse cohorts.
Keywords: KCNMB2; Dementia; Diversity; Epidemiology; FTLD; Genome-Wide Association Studies (GWAS); KATP.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Association of Neuropathologists, Inc.
Conflict of interest statement
The authors have no duality or conflicts of interest to declare.
Figures



References
-
- So HC, Gui AH, Cherny SS, et al. Evaluating the heritability explained by known susceptibility variants: A survey of ten complex diseases. Genet Epidemiol 2011;35:310–7 - PubMed
-
- Kovacs GG, Milenkovic I, Wohrer A, et al. Non-Alzheimer neurodegenerative pathologies and their combinations are more frequent than commonly believed in the elderly brain: A community-based autopsy series. Acta Neuropathol 2013;126:365–84 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- P50 AG023501/AG/NIA NIH HHS/United States
- R01 AG022374/AG/NIA NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
- R01 AG042437/AG/NIA NIH HHS/United States
- R01 AG031581/AG/NIA NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- P50 MH060451/MH/NIMH NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 AG008671/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- U54 AG052427/AG/NIA NIH HHS/United States
- RC2 AG036528/AG/NIA NIH HHS/United States
- P30 AG028377/AG/NIA NIH HHS/United States
- R01 AG013616/AG/NIA NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- R01 AG061111/AG/NIA NIH HHS/United States
- R01 AG009029/AG/NIA NIH HHS/United States
- R01 AG030146/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- KL2 RR024151/RR/NCRR NIH HHS/United States
- P50 AG005128/AG/NIA NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- P50 AG033514/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- U01 HG008657/HG/NHGRI NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- RC2 AG036502/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- P20 MD000546/MD/NIMHD NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- R01 AG020688/AG/NIA NIH HHS/United States
- RF1 NS118584/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- F30 NS124136/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P50 NS039764/NS/NINDS NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- R01 AG009956/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- R01 MH080295/MH/NIMH NIH HHS/United States
- R01 AG026390/AG/NIA NIH HHS/United States
- R01 AG028786/AG/NIA NIH HHS/United States
- U19 AG024904/AG/NIA NIH HHS/United States
- R01 AG015473/AG/NIA NIH HHS/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- U24 AG041689/AG/NIA NIH HHS/United States
- R01 AG012101/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- P30 AG008017/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- UL1 RR029893/RR/NCRR NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R01 AG019085/AG/NIA NIH HHS/United States
- P50 AG016570/AG/NIA NIH HHS/United States
- R37 AG015473/AG/NIA NIH HHS/United States
- F30 NS124136/NS/NINDS NIH HHS/United States
- U24 AG026395/AG/NIA NIH HHS/United States
- R01 AG035137/AG/NIA NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- P01 AG019724/AG/NIA NIH HHS/United States
- R01 CA129769/CA/NCI NIH HHS/United States
- U01 AG010483/AG/NIA NIH HHS/United States
- P30 AG028383/AG/NIA NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- R01 AG027944/AG/NIA NIH HHS/United States
- R01 AG041232/AG/NIA NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- R01 AG048927/AG/NIA NIH HHS/United States
- R01 AG037212/AG/NIA NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- R01 AG017173/AG/NIA NIH HHS/United States
- R01 AG025259/AG/NIA NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- RF1 AG015473/AG/NIA NIH HHS/United States
- U01 HG004610/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P30 AG019610/AG/NIA NIH HHS/United States
- RF1 AG082339/AG/NIA NIH HHS/United States
- RC2 AG036650/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 AG026916/AG/NIA NIH HHS/United States
- P50 AG016582/AG/NIA NIH HHS/United States
- R01 NS059873/NS/NINDS NIH HHS/United States

