A genetically encoded sensor for visualizing leukotriene B4 gradients in vivo
- PMID: 37528073
- PMCID: PMC10393954
- DOI: 10.1038/s41467-023-40326-6
A genetically encoded sensor for visualizing leukotriene B4 gradients in vivo
Abstract
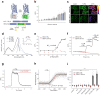
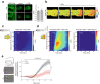
Leukotriene B4 (LTB4) is a potent lipid chemoattractant driving inflammatory responses during host defense, allergy, autoimmune and metabolic diseases. Gradients of LTB4 orchestrate leukocyte recruitment and swarming to sites of tissue damage and infection. How LTB4 gradients form and spread in live tissues to regulate these processes remains largely elusive due to the lack of suitable tools for monitoring LTB4 levels in vivo. Here, we develop GEM-LTB4, a genetically encoded green fluorescent LTB4 biosensor based on the human G-protein-coupled receptor BLT1. GEM-LTB4 shows high sensitivity, specificity and a robust fluorescence increase in response to LTB4 without affecting downstream signaling pathways. We use GEM-LTB4 to measure ex vivo LTB4 production of murine neutrophils. Transgenic expression of GEM-LTB4 in zebrafish allows the real-time visualization of both exogenously applied and endogenously produced LTB4 gradients. GEM-LTB4 thus serves as a broadly applicable tool for analyzing LTB4 dynamics in various experimental systems and model organisms.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

