Common antibiotics, azithromycin and amoxicillin, affect gut metagenomics within a household
- PMID: 37528343
- PMCID: PMC10394940
- DOI: 10.1186/s12866-023-02949-z
Common antibiotics, azithromycin and amoxicillin, affect gut metagenomics within a household
Abstract
Background: The microbiome of the human gut serves a role in a number of physiological processes, but can be altered through effects of age, diet, and disturbances such as antibiotics. Several studies have demonstrated that commonly used antibiotics can have sustained impacts on the diversity and the composition of the gut microbiome. The impact of the two most overused antibiotics, azithromycin, and amoxicillin, in the human microbiome has not been thoroughly described. In this study, we recruited a group of individuals and unrelated controls to decipher the effects of the commonly used antibiotics amoxicillin and azithromycin on their gut microbiomes.
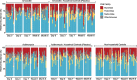
Results: We characterized the gut microbiomes by metagenomic sequencing followed by characterization of the resulting microbial communities. We found that there were clear and sustained effects of the antibiotics on the gut microbial community with significant alterations in the representations of Bifidobacterium species in response to azithromycin (macrolide antibiotic). These results were supported by significant increases identified in putative antibiotic resistance genes associated with macrolide resistance. Importantly, we did not identify these trends in the unrelated control individuals. There were no significant changes observed in other members of the microbial community.
Conclusions: As we continue to focus on the role that the gut microbiome plays and how disturbances induced by antibiotics might affect our overall health, elucidating members of the community most affected by their use is of critical importance to understanding the impacts of common antibiotics on those who take them. Clinical Trial Registration Number NCT05169255. This trial was retrospectively registered on 23-12-2021.
Keywords: Antibiotic courses; Antibiotic perturbations; Bacteriophage; Beta Lactam; Fecal; Gut; Macrolide; Metagenome; Microbiome; Microbiota; Resistome; Virome; Virus.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

