Federated generalized linear mixed models for collaborative genome-wide association studies
- PMID: 37529100
- PMCID: PMC10387571
- DOI: 10.1016/j.isci.2023.107227
Federated generalized linear mixed models for collaborative genome-wide association studies
Abstract
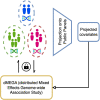
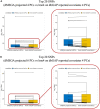
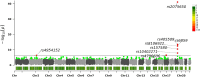
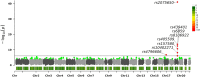
Federated association testing is a powerful approach to conduct large-scale association studies where sites share intermediate statistics through a central server. There are, however, several standing challenges. Confounding factors like population stratification should be carefully modeled across sites. In addition, it is crucial to consider disease etiology using flexible models to prevent biases. Privacy protections for participants pose another significant challenge. Here, we propose distributed Mixed Effects Genome-wide Association study (dMEGA), a method that enables federated generalized linear mixed model-based association testing across multiple sites without explicitly sharing genotype and phenotype data. dMEGA employs a reference projection to correct for population-stratification and utilizes efficient local-gradient updates among sites, incorporating both fixed and random effects. The accuracy and efficiency of dMEGA are demonstrated through simulated and real datasets. dMEGA is publicly available at https://github.com/Li-Wentao/dMEGA.
Keywords: Clinical genetics; Genomics; Health sciences; Human genetics.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Palsson G., Rabinow P. Iceland: the case of a national human genome project. Anthropol. Today. 1999;15:14–18. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

