Towards design of drugs and delivery systems with the Martini coarse-grained model
- PMID: 37529288
- PMCID: PMC10392664
- DOI: 10.1017/qrd.2022.16
Towards design of drugs and delivery systems with the Martini coarse-grained model
Abstract
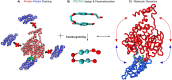
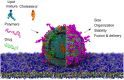
Coarse-grained (CG) modelling with the Martini force field has come of age. By combining a variety of bead types and sizes with a new mapping approach, the newest version of the model is able to accurately simulate large biomolecular complexes at millisecond timescales. In this perspective, we discuss possible applications of the Martini 3 model in drug discovery and development pipelines and highlight areas for future development. Owing to its high simulation efficiency and extended chemical space, Martini 3 has great potential in the area of drug design and delivery. However, several aspects of the model should be improved before Martini 3 CG simulations can be routinely employed in academic and industrial settings. These include the development of automatic parameterisation protocols for a variety of molecule types, the improvement of backmapping procedures, the description of protein flexibility and the development of methodologies enabling efficient sampling. We illustrate our view with examples on key areas where Martini could give important contributions such as drugs targeting membrane proteins, cryptic pockets and protein-protein interactions and the development of soft drug delivery systems.
Keywords: Martini; PROTACS; coarse-grained models; cryptic pockets; drug delivery; drug design; lipid nanoparticles; molecular dynamics; protein-protein interactions; soft delivery systems; transmembrane proteins.
© The Author(s) 2022.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
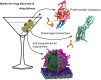

References
-
- Alessandri R, Barnoud J, Gertsen AS, Patmanidis I, de Vries AH, Souza PCT and Marrink SJ (2022) Martini 3 coarse-grained force field: Small molecules. Advanced Theory and Simulations 5, 2100391.
