Investigating the composition and recruitment of the mycobacterial ImuA'-ImuB-DnaE2 mutasome
- PMID: 37530405
- PMCID: PMC10421592
- DOI: 10.7554/eLife.75628
Investigating the composition and recruitment of the mycobacterial ImuA'-ImuB-DnaE2 mutasome
Abstract
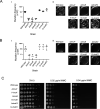
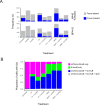
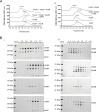
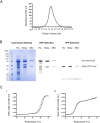
A DNA damage-inducible mutagenic gene cassette has been implicated in the emergence of drug resistance in Mycobacterium tuberculosis during anti-tuberculosis (TB) chemotherapy. However, the molecular composition and operation of the encoded 'mycobacterial mutasome' - minimally comprising DnaE2 polymerase and ImuA' and ImuB accessory proteins - remain elusive. Following exposure of mycobacteria to DNA damaging agents, we observe that DnaE2 and ImuB co-localize with the DNA polymerase III β subunit (β clamp) in distinct intracellular foci. Notably, genetic inactivation of the mutasome in an imuBAAAAGG mutant containing a disrupted β clamp-binding motif abolishes ImuB-β clamp focus formation, a phenotype recapitulated pharmacologically by treating bacilli with griselimycin and in biochemical assays in which this β clamp-binding antibiotic collapses pre-formed ImuB-β clamp complexes. These observations establish the essentiality of the ImuB-β clamp interaction for mutagenic DNA repair in mycobacteria, identifying the mutasome as target for adjunctive therapeutics designed to protect anti-TB drugs against emerging resistance.
Keywords: Mycobacterium smegmatis; Mycobacterium tuberculosis; anti-evolution; antibiotic resistance; biochemistry; chemical biology; induced mutagenesis; infectious disease; microbiology; mutasome.
Conflict of interest statement
SG, ZM, MR, JS, RD, AR, ND, Td, SA, DL, JA, TC, JH, RM, JM, RW, VM, ČV, ML, DW No competing interests declared
Figures






Update of
References
-
- Bosch B, DeJesus MA, Poulton NC, Zhang W, Engelhart CA, Zaveri A, Lavalette S, Ruecker N, Trujillo C, Wallach JB, Li S, Ehrt S, Chait BT, Schnappinger D, Rock JM. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell. 2021;184:4579–4592. doi: 10.1016/j.cell.2021.06.033. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

