A modular plasmid toolkit applied in marine bacteria reveals functional insights during bacteria-stimulated metamorphosis
- PMID: 37530556
- PMCID: PMC10470607
- DOI: 10.1128/mbio.01502-23
A modular plasmid toolkit applied in marine bacteria reveals functional insights during bacteria-stimulated metamorphosis
Abstract
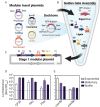
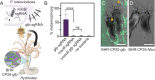
A conspicuous roadblock to studying marine bacteria for fundamental research and biotechnology is a lack of modular synthetic biology tools for their genetic manipulation. Here, we applied, and generated new parts for, a modular plasmid toolkit to study marine bacteria in the context of symbioses and host-microbe interactions. To demonstrate the utility of this plasmid system, we genetically manipulated the marine bacterium Pseudoalteromonas luteoviolacea, which stimulates the metamorphosis of the model tubeworm, Hydroides elegans. Using these tools, we quantified constitutive and native promoter expression, developed reporter strains that enable the imaging of host-bacteria interactions, and used CRISPR interference (CRISPRi) to knock down a secondary metabolite and a host-associated gene. We demonstrate the broader utility of this modular system for testing the genetic tractability of marine bacteria that are known to be associated with diverse host-microbe symbioses. These efforts resulted in the successful conjugation of 12 marine strains from the Alphaproteobacteria and Gammaproteobacteria classes. Altogether, the present study demonstrates how synthetic biology strategies enable the investigation of marine microbes and marine host-microbe symbioses with potential implications for environmental restoration and biotechnology. IMPORTANCE Marine Proteobacteria are attractive targets for genetic engineering due to their ability to produce a diversity of bioactive metabolites and their involvement in host-microbe symbioses. Modular cloning toolkits have become a standard for engineering model microbes, such as Escherichia coli, because they enable innumerable mix-and-match DNA assembly and engineering options. However, such modular tools have not yet been applied to most marine bacterial species. In this work, we adapt a modular plasmid toolkit for use in a set of 12 marine bacteria from the Gammaproteobacteria and Alphaproteobacteria classes. We demonstrate the utility of this genetic toolkit by engineering a marine Pseudoalteromonas bacterium to study their association with its host animal Hydroides elegans. This work provides a proof of concept that modular genetic tools can be applied to diverse marine bacteria to address basic science questions and for biotechnology innovations.
Keywords: CRISPRi; Hydroides; golden gate; marine; metamorphosis; modular; symbiosis; tubeworm; violacein.
Conflict of interest statement
A.T.A. and N.J.S. are coinventors on provisional U.S. patent application serial number 63/323653, entitled "Genetic Engineering of Marine Bacteria for Biomaterial Production, Probiotic Use in Aquaculture and Marine Environmental Restoration" and assigned to San Diego State University Research Foundation.
Figures


Update of
-
A modular plasmid toolkit applied in marine Proteobacteria reveals functional insights during bacteria-stimulated metamorphosis.bioRxiv [Preprint]. 2023 Jan 31:2023.01.31.526474. doi: 10.1101/2023.01.31.526474. bioRxiv. 2023. Update in: mBio. 2023 Aug 31;14(4):e0150223. doi: 10.1128/mbio.01502-23. PMID: 36778221 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

