Vitamin D induces SIRT1 activation through K610 deacetylation in colon cancer
- PMID: 37530744
- PMCID: PMC10396337
- DOI: 10.7554/eLife.86913
Vitamin D induces SIRT1 activation through K610 deacetylation in colon cancer
Abstract
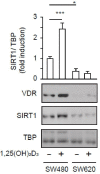
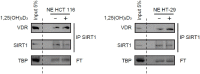
Posttranslational modifications of epigenetic modifiers provide a flexible and timely mechanism for rapid adaptations to the dynamic environment of cancer cells. SIRT1 is an NAD+-dependent epigenetic modifier whose activity is classically associated with healthy aging and longevity, but its function in cancer is not well understood. Here, we reveal that 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3, calcitriol), the active metabolite of vitamin D (VD), promotes SIRT1 activation through auto-deacetylation in human colon carcinoma cells, and identify lysine 610 as an essential driver of SIRT1 activity. Remarkably, our data show that the post-translational control of SIRT1 activity mediates the antiproliferative action of 1,25(OH)2D3. This effect is reproduced by the SIRT1 activator SRT1720, suggesting that SIRT1 activators may offer new therapeutic possibilities for colon cancer patients who are VD deficient or unresponsive. Moreover, this might be extrapolated to inflammation and other VD deficiency-associated and highly prevalent diseases in which SIRT1 plays a prominent role.
Keywords: VDR; acetylation; cancer biology; colorrectal cancer; human; vitamin D.
© 2023, García-Martínez et al.
Conflict of interest statement
JG, AC, JM, MF, MF, JC, AD, AB, AM, ML, CG No competing interests declared
Figures






Update of
- doi: 10.1101/2023.02.22.529558
- doi: 10.7554/eLife.86913.1
- doi: 10.7554/eLife.86913.2
References
-
- Bartha Á, Győrffy B. TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. International Journal of Molecular Sciences. 2021;22:1–12. doi: 10.3390/ijms22052622. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

