Differential Expression of miRNAs in Amyotrophic Lateral Sclerosis Patients
- PMID: 37531027
- PMCID: PMC10657797
- DOI: 10.1007/s12035-023-03520-7
Differential Expression of miRNAs in Amyotrophic Lateral Sclerosis Patients
Abstract
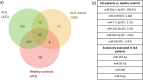
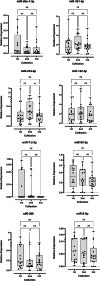
Amyotrophic lateral sclerosis (ALS) is a progressive motor neuron disease that affects nerve cells in the brain and spinal cord, causing loss of muscle control, muscle atrophy and in later stages, death. Diagnosis has an average delay of 1 year after symptoms onset, which impairs early management. The identification of a specific disease biomarker could help decrease the diagnostic delay. MicroRNA (miRNA) expression levels have been proposed as ALS biomarkers, and altered function has been reported in ALS pathogenesis. The aim of this study was to assess the differential expression of plasma miRNAs in ALS patients and two control populations (healthy controls and ALS-mimic disorders). For that, 16 samples from each group were pooled, and then 1008 miRNAs were assessed through reverse transcription-quantitative polymerase chain reaction (RT-qPCR). From these, ten candidate miRNAs were selected and validated in 35 ALS patients, 16 ALS-mimic disorders controls and 15 healthy controls. We also assessed the same miRNAs in two different time points of disease progression. Although we were unable to determine a miRNA signature to use as disease or condition marker, we found that miR-7-2-3p, miR-26a-1-3p, miR-224-5p and miR-206 are good study candidates to understand the pathophysiology of ALS.
Keywords: Amyotrophic lateral sclerosis; Biomarkers; Epigenetics; MicroRNAs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

